What is Pharmacogenomics?
Pharmacogenomics
Pharmacogenomics represents a cutting-edge field merging molecular pharmacology and functional genomics, emerging in the late 1990s. Its core objective lies in enhancing drug efficacy and safety while dissecting the impact of genetic variations on individual responses to medications, thus refining the rational utilization of pharmaceuticals.
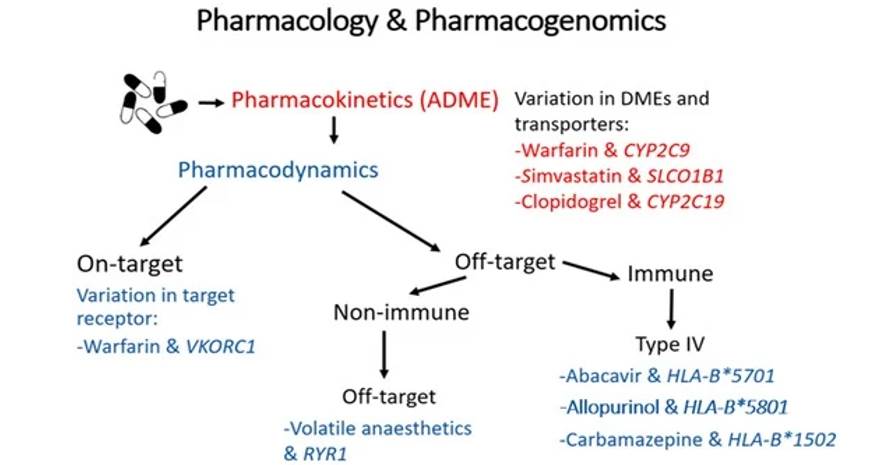 A summary of the mechanisms by which genomic variation can influence the pharmacokinetics and/or pharmacodynamics of a drug, with examples of clinically relevant drug-gene pairs. (Rollinson et al., 2020)
A summary of the mechanisms by which genomic variation can influence the pharmacokinetics and/or pharmacodynamics of a drug, with examples of clinically relevant drug-gene pairs. (Rollinson et al., 2020)
The History of Pharmacogenomics
Pharmacogenomics traces its origins back to 1957 when Arno Motulsky of the University of Washington and Werner Kalow of the University of Toronto postulated that genetic variations in enzyme activity among individuals could lead to diverse or even contrasting responses to drugs. However, it wasn't until 1997 that significant strides were made in understanding the role of cytochrome P450 enzymes (CYP450s) in drug metabolism. An associate professor of pharmacy at the University of Washington discovered that, for most drugs, the activity of CYP450s determines both the duration and concentration of drug presence in the body. Six distinct cytochromes—CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4—play crucial roles in metabolizing drugs, with notable correlations identified between polymorphisms in CYP2C9, CYP2C19, and CYP2D6 and inter-individual variations.
In 1997, Genset and Abbott Laboratories (Abbott Park, IL) forged an agreement to jointly develop and promote genetic diagnostic kits aimed at assessing drug efficacy in patients, marking a pivotal moment in the advancement of pharmacogenomics. The term "pharmacogenomics" itself was introduced in 1999, marking the formal recognition of this burgeoning field. Since then, ongoing research has progressively elucidated the intricate relationship between genetic polymorphisms and individual variations in drug response, underscoring the transformative potential of pharmacogenomics in personalized medicine.
Applications of Pharmacogenomics
Pharmacogenomics delves into the intricate interplay between human genomic information and drug responses, aiming to decipher the underlying causes of individualized variations in drug action. By harnessing genomic insights, pharmacogenomics endeavors to pave the way for the development of novel drugs and the realization of tailored medication regimens tailored to individual needs. These variations encompass a spectrum of factors, including individual drug sensitivity, metabolic rates, and susceptibility to adverse reactions or toxicity.
Key areas of focus within pharmacogenomics include:
- Predicting Drug Sensitivity: Through the assessment of genetic markers, pharmacogenomics endeavors to forecast an individual's sensitivity to particular drugs, thereby enhancing the prediction of drug efficacy.
- Predicting Metabolic Rate: By examining the role of metabolizing enzymes and transporters encoded by genetic variations, pharmacogenomics aims to predict the rate at which a drug is metabolized within the body. This understanding aids in optimizing dosage regimens tailored to individual metabolic profiles.
- Anticipating Adverse Drug Reactions: Pharmacogenomics seeks to identify genetic markers associated with adverse drug reactions, facilitating the avoidance or mitigation of potential drug-related complications.
In essence, pharmacogenomics holds the promise of revolutionizing the field of medicine by ushering in an era of personalized therapeutics, where treatments are precisely tailored to individual genetic makeup, thereby maximizing efficacy while minimizing adverse effects.
Cutting-edge technologies, such as high-throughput sequencing and long-read sequencing, employed by CD Genomics, facilitate the robust analysis of gene mutations and drug reactions, based on genome information. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
Pharmacogenomics: Understanding its Impact on Drug Effects
The absorption, distribution, metabolism, and excretion of drugs within the human body are intricately linked to the body's physiological mechanisms, governed by a complex interplay of genes encoding drug-metabolizing enzymes, transporters, receptors, and other proteins. Genetic mutations can augment, weaken, or abolish protein function, leading to alterations in drug action processes or changes in drug-target binding affinity, ultimately influencing drug efficacy or triggering adverse effects. At the core of these effects lie gene polymorphisms, commonly referred to as single nucleotide polymorphisms (SNPs).
The Human Genome Project has revealed that individuals' gene sequences differ by only one in a thousand, resulting in approximately 3 million variants. These variants, or polymorphisms, occur at a rate of approximately one variant per 500-1000 bases in the genome, giving rise to distinct genotypes that dictate individuals' susceptibility to diseases and their unique responses to drugs.
SNPs, representing variations in a single nucleotide within the DNA sequence, occur at a frequency exceeding 1% in the population. When a nucleotide, such as A, is substituted by any of the other three nucleotides (C/T/G), it gives rise to SNPs, representing the smallest genetic differences among individuals. Remarkably, 90% of human genetic diversity is attributed to SNPs, with pharmacogenomics extensively analyzing inter-individual drug differences from an SNP perspective.
Pharmacogenomic research predominantly focuses on four categories of drug-related genes: (1) enzymes involved in drug metabolism pathways, (2) receptors (target proteins) that interact with drugs, such as ADRB2, (3) drug-transporter channels, such as the MDR1 gene, and (4) signaling-related proteins. It's worth noting that a significant proportion—59%—of adverse drug reactions stem from genetic polymorphisms in drug-metabolizing enzymes.
Technologies of Pharmacogenomics
With advancements in genomics and the implementation of various genome-based initiatives, the feasibility of pharmacogenetic testing has been firmly established through the design of probes targeting drug-related genes and subsequent clinical validation. The identification of genetic variations encompasses numerous methods, broadly categorized into two main approaches.
The first category comprises traditional methods reliant on gel electrophoresis, including restriction fragment length polymorphism (PCR-RFLP), single-stranded conformation polymorphism (PCR-SSCP), denaturing gradient gel electrophoresis (PCR-DGGE), and allele-specific PCR (AS-PCR). These methods have long been employed in genetic analysis but may be labor-intensive and time-consuming.
In contrast, the second category embraces high-throughput sequencing and automation, representing more recent developments in genetic analysis. Techniques such as Sanger sequencing, DNA microarray detection, next-generation sequencing, denaturing high-performance liquid chromatography (DHPLC), and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) offer increased throughput and automation, streamlining the genetic analysis process. These advanced methods enable rapid and efficient screening of genetic variations, facilitating the widespread adoption of pharmacogenetic testing in clinical practice.
From Reactive to Preemptive Testing
Reactive testing poses significant drawbacks, primarily centered on decision-making delays inherent in the testing process. This delay can lead to inappropriate treatment choices, potentially resulting in medication toxicity or inefficacy. For instance, the administration of clopidogrel in emergency PCI patients without prior knowledge of their CYP2C19 genotype underscores this issue. Conversely, preemptive testing ensures that pharmacogenetic test results are readily available in the patient's medical record prior to prescription. This proactive approach encompasses both single-gene testing and, more prominently, panel or whole-exome testing, encompassing multiple actionable pharmacogenetic genes.
Compared to reactive single-gene testing, preemptive multigene panel testing offers several advantages. Firstly, it streamlines efficiency by obviating the need for multiple patient samples and DNA isolations, as a single pharmacogenetic panel suffices. Secondly, it eliminates therapeutic delays as test results are already integrated into the patient's medical record at the point of prescription. Notably, preemptive polygenic panel testing further expedites treatment decisions by preemptively embedding test results in patient records, negating the need to await result returns. Importantly, pharmacogenomic testing is typically a one-time endeavor, with individual results retrievable as needed in subsequent medical scenarios.
Currently, the adoption of NGS technology in pharmacogenomics testing remains limited due to cost, technological complexity, and result interpretation challenges. However, the increasing accessibility and utilization of high-throughput sequencing portend a future where pharmacogenetic testing, particularly preemptive testing, becomes commonplace.
Genotyping Techniques: PCR-RFLP
Polymerase chain reaction/restriction endonuclease cleaved fragment length polymorphism (PCR-RFLP) analysis is commonly employed for genotyping prevalent SNPs within genes. This method, albeit still utilized in research contexts, presents challenges in terms of complexity and labor intensiveness, rendering it unsuitable for routine clinical diagnostics or high-throughput applications. Moreover, due to the necessity of manipulating reaction tubes post-PCR and treating primers with restriction enzymes, there's a heightened risk of PCR cross-contamination. As a precaution, physical segregation between sample preparation, PCR setup, and post-PCR analysis is imperative to prevent false-positive outcomes arising from contamination.
- Principles of PCR-RFLP
While certain sequence variants, such as single base changes, may not alter DNA length, they can create or disrupt recognition sites for endonucleases. PCR amplifies the target sequence, after which the amplification product undergoes digestion with restriction endonucleases to ascertain changes in the restriction site. The status of the site can be discerned through gel electrophoresis of the digestion product. For instance, a sample harboring a variant that disrupts the enzyme recognition site will yield an uncut PCR fragment, unlike a sample lacking the variant, which produces two shorter fragments. In cases of heterozygosity (presence of one normal and one variant allele), one long fragment and two shorter fragments are observed. This assay can be designed to facilitate easy differentiation of fragments via agarose electrophoresis.
Allele-specific PCR (AS-PCR)
Allele-specific PCR (AS-PCR) capitalizes on the disparate amplification efficiencies of two homologous primers differing solely at their terminal 3' positions, with each primer precisely complementing either the normal or variant allele. Analysis of the reaction products typically employs gel-based methods or, more commonly, homogeneous real-time PCR utilizing fluorescent dyes. The latter is favored for its convenience over electrophoresis. Generally, fluorescent dye-based AS-PCR necessitates at least two PCR reactions per polymorphism analysis. Moreover, AS-PCR hinges on optimizing PCR selectivity to ensure that the fluorescent signal, resulting from dye binding to the amplicon, accurately reflects the expected amplification product, free from interference from undesired side reactions like primer dimerization.
Given the preference for direct polymorphism detection in clinical diagnostics and the often requisite detection of multiple polymorphisms in genotyping assays, various multiplex techniques have emerged to address this demand. These include leveraging TaqMan probes, hybridization probes, and Invader probes.
TaqMan Probes
TaqMan probes are widely utilized in both single and multi-locus genotyping methodologies, employing a probe-based homogeneous PCR approach for direct variant detection. Among the diverse variant detection methods, the predominant choice involves 5' nuclease hydrolysis probes, commonly known as TaqMan probes. These probes, distinctly labeled with fluorescent markers, are meticulously designed to perfectly complement either of the two variant alleles differing by one or more nucleotides.
Functionally, these doubly labeled probes are initially quenched due to their proximity in the oligonucleotide probe conformation. During each PCR cycle, the TaqMan probe hybridizes with its target region, and at the primer extension step, the fluorescent reporter fluorophore and the quenched fluorophore dissociate, emitting fluorescence upon cleavage by the 5' nuclease activity of a heat-resistant DNA polymerase (e.g., Thermus aquaticus). Consequently, the fluorescence intensity, associated with each allele-specific TaqMan probe, steadily increases in real-time with each subsequent amplification cycle.
The detection of both alleles, whether in a pure or heterozygous state, is typically determined by cycles in which the observable signal surpasses a preset threshold (denoted as CT). However, the ability to genotype multiple SNPs within a single PCR reaction is constrained more by the number of distinct fluorescent signals detectable in a single reaction than by the multiplexing capacity to target several regions concurrently. This limitation hinges on factors such as the emission spectra overlap of fluorescent dyes used for probe labeling, detection hardware capabilities, and cross-talk correction software efficacy.
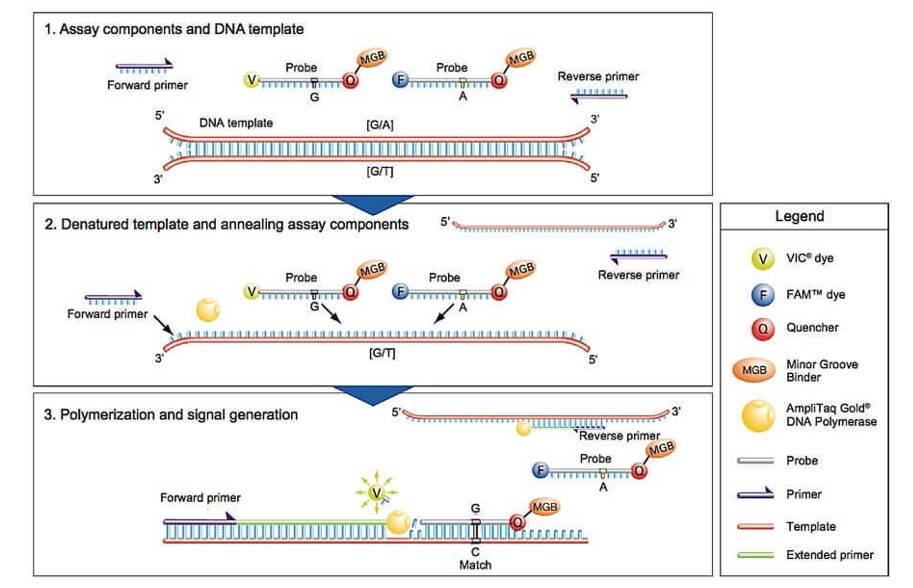 TaqMan genotyping platform
TaqMan genotyping platform
Hybridization Probes
Hybridization probes present a robust method for homogeneous fluorescent genotyping, particularly through post-PCR melting curve analysis of allele-specific hybridization probes. In this approach, two probes are strategically designed for each SNP site: the anchoring probe selectively binds to sequences near the SNP site, while the SNP detection probe specifically recognizes the SNP-containing sequence. Leveraging the fluorescence resonance energy transfer (FRET) principle, the anchoring probe is labeled with a donor fluorescent dye at one end, while the SNP detection probe carries an acceptor dye at one end. The probes are designed to optimally hybridize at a temperature conducive to both binding adjacent to each other on the PCR product, facilitating fluorescent signal generation during and after PCR.
SNP detection relies on melting curve analysis, a technique that discriminates between different allelic forms of DNA based on their melting temperatures. Specifically, SNP detection probes are engineered to exhibit melting temperatures distinct from those of wild-type and variant SNPs. As the temperature fluctuates, the probe dissociates from the target sequence at a specific melting temperature, contingent upon the allelic composition of the SNP. The resulting melting profiles, unique to each SNP, enable the detection and characterization of SNP types.
Gene Microarray
In hybridization-based techniques, the process initiates with the isolation of genomic DNA, followed by PCR amplification of the region of interest. Subsequently, the amplified target is fragmented using DNase I enzyme and end-labeled with biotin. This labeled product is then subjected to hybridization with allele-specific oligonucleotides on a solid substrate, such as beads or microarrays, under stringent conditions. These allele-specific oligonucleotides, differing by only one or two bases, correspond to the various alleles of the DNA fragment under examination. The reaction environment is carefully controlled to allow the removal of mismatched targets, ensuring that only perfectly hybridized DNA fragments remain, which are then fluorescently labeled. The fluorescence associated with specific probe features is detected using a laser-illuminated confocal scanner.
To enhance the specificity of the assay, multiple probes are often employed for each SNP allele. Oligonucleotide array-based assays typically leverage comparative fluorescence signals between sets of perfectly matched and mismatched oligonucleotides to detect polymorphisms while mitigating the effects of cross-hybridization. Identification of polymorphisms, alleles, and genotypes within a sample is facilitated by training software and algorithms, utilizing samples with known genotypes. For instance, this methodology has been applied in the development of the AmpliChip CYP450 genotyping assay.
The gene microarray method enables the simultaneous detection of nearly all known polymorphisms and alleles of CYP2D6 via multiplexed long PCR reactions. These reactions amplify the promoter and coding region of CYP2D6, along with specific reactions tailored for detecting a 3.5 kb CYP2D6 gene deletion or a CYP2D6 gene duplication.
Sanger Sequencing
Sanger sequencing revolutionized genetic analysis when introduced by Sanger et al. in 1977. Over the years, this technology has undergone significant refinement and automation, culminating in today's advanced Sanger sequencing platforms. Modern Sanger sequencing utilizes four-color fluorescent dyes for ddNTP labeling, coupled with capillary electrophoresis to precisely separate DNA fragments. These advancements have substantially enhanced the convenience, safety, and throughput of sequencing procedures.
Renowned as a classic method for DNA sequence analysis, Sanger sequencing is widely regarded as the gold standard for genotyping due to its ability to directly read DNA sequences. This versatility makes it suitable for detecting various genetic variations, including single nucleotide polymorphisms (SNPs), short tandem repeats (STRs), Alu repeat sequence polymorphisms (ARPs), and HLA-B typing.
Notably, several class III in vitro diagnostic (IVD) reagents based on Sanger sequencing have received approval from regulatory agencies like the National Medical Products Administration (NMPA). These reagents facilitate the detection of genetic variations at loci such as UGT1A1, ALDH2, MTHFR, CYP2C19, CYP2C9, VKORC1, and others, underscoring the widespread applicability and clinical utility of Sanger sequencing in molecular diagnostics.
Next-Generation Sequencing (NGS)
High-throughput sequencing, also known as next-generation sequencing (NGS) or massively parallel sequencing (MPS), represents a groundbreaking leap forward from Sanger sequencing. This innovative approach capitalizes on parallel sequencing, enabling the simultaneous sequencing of millions or even billions of DNA fragments, thereby achieving large-scale, high-throughput sequencing objectives. Prominent commercialized NGS technologies, such as Illumina's Solexa technology and Thermo's Ion Torrent technology, dominate the landscape of high-throughput sequencing platforms.
The primary advantage of high-throughput sequencing lies in its ability to achieve remarkable throughput while concurrently detecting various genetic variations, including single nucleotide polymorphisms (SNPs), copy number variations (CNVs), short tandem repeats (STRs), Alu repeats, and HLA-B typing. However, the method also presents several challenges. The instrumentation is costly, and the reagent expenses can be substantial. Additionally, the complexity of the detection process, coupled with stringent requirements for laboratory facilities and skilled operators, poses practical hurdles. Furthermore, data analysis can be intricate, impeding widespread adoption.
High-throughput sequencing finds particular utility in applications involving the detection of genetic variants across multiple gene regions or the identification of mutations present at low frequencies. Despite its complexities and limitations, high-throughput sequencing stands at the forefront of genetic analysis, driving advancements in molecular diagnostics and research endeavors.
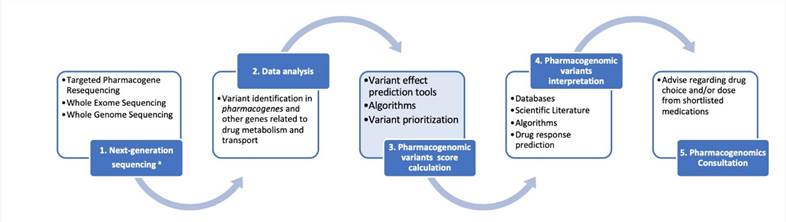 A schematic representation for the clinical pharmacogenomics workflow described herein. (Giannopoulou et al., 2019)
A schematic representation for the clinical pharmacogenomics workflow described herein. (Giannopoulou et al., 2019)
Cutting-edge technologies, such as high-throughput sequencing and long-read sequencing, employed by CD Genomics, facilitate the robust analysis of gene mutations and drug reactions, based on genome information. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
In Summary
In recent years, the emergence of multi-omics approaches has revolutionized our ability to forecast diseases and predict drug responses, facilitating precise diagnosis and treatment strategies. Among these multi-omics technologies, pharmacogenomics stands out by elucidating the intricate interplay between genetic information and drug pharmacokinetics/pharmacodynamics (PK/PD) through DNA sequencing data.
When integrated into genome-wide association studies, transcriptomics, proteomics, epigenetics, and metabolomics hold tremendous potential for uncovering genetic factors predisposing individuals to multifactorial diseases and varied drug responses. This comprehensive approach enables the early identification of susceptible populations, paving the way for targeted interventions aimed at enhancing drug safety, efficacy, and mitigating adverse effects.
Moreover, the fusion of proteomics and metabolomics further enhances our ability to discern genetic factors associated with multifactorial diseases and drug reactions. By enabling early interventions tailored to susceptible populations, this integrative approach promotes optimized drug therapies that maximize efficacy while minimizing toxicity. Ultimately, it propels us towards the realization of safe, effective, and economically viable individualized drug treatments, thereby fostering enhanced human health and well-being.
References:
- Rollinson, Victoria, Richard Turner, and Munir Pirmohamed. "Pharmacogenomics for primary care: an overview." Genes 11.11 (2020): 1337.
- Giannopoulou, Efstathia, et al. "Integrating next-generation sequencing in the clinical pharmacogenomics workflow." Frontiers in pharmacology 10 (2019): 384.


 Sample Submission Guidelines
Sample Submission Guidelines
