How to Plan Your Next RNA Sequencing Experiment
What is RNA sequencing?
RNA-Seq, an immensely sensitive and precise technique, has emerged as an indispensable tool for investigating gene expression and has facilitated significant breakthroughs across numerous scientific disciplines. While conventional methods like microarray microarrays solely measure known genes and transcripts, they fall short in capturing novel features and unidentified transcripts. Conversely, RNA-Seq leverages high-throughput sequencing technology to convert RNA samples into quantifiable DNA sequences, thus enabling researchers to detect both established and novel features unencumbered by prior knowledge constraints.
One of the distinct advantages of RNA-Seq is its capacity to identify heterodimers of transcripts, namely different splice variants originating from the same gene, which may exhibit diverse expression patterns across various tissues, developmental stages, or disease states. Moreover, RNA-Seq enables the detection of gene fusion events, a phenomenon commonly observed in certain tumors where two or more genes fuse together. Additionally, RNA-Seq plays a pivotal role in detecting single nucleotide variants (SNVs), which denote variations in a single nucleotide within the genome. Such variants assume critical significance in the examination of genetic disparities between individuals and in identifying genes associated with diseases.
By providing transcriptome-wide expression information, RNA-Seq empowers researchers to unravel intricate gene regulatory mechanisms, comprehend cellular function, and elucidate the progression of diseases. Furthermore, RNA-Seq facilitates comparative analyses of gene expression disparities under distinct conditions, such as responses to therapeutic drugs and adaptations to diverse environmental contexts.
Please read our article RNA-Seq for Differential Gene Expression Analysis: Introduction, Protocol, and Bioinformatics for more information.
Benefits of RNA sequencing
RNA sequencing (RNA-Seq) with next-generation sequencing (NGS) technology offers several benefits for studying the transcriptome:
- Broad dynamic range: RNA-Seq can detect and quantify gene expression across a wide range of expression levels, from highly abundant transcripts to rare transcripts. This allows researchers to capture subtle changes in gene expression and identify lowly expressed genes that may have important biological functions.
- Sensitive and accurate measurement: RNA-Seq provides a high level of sensitivity and accuracy in measuring gene expression. It can detect even small changes in gene expression levels, making it suitable for various applications, including differential gene expression analysis and identifying differentially expressed genes under different conditions.
- Detection of known and novel features: RNA-Seq allows for the detection of both known and novel features without prior knowledge or the need for predesigned probes. It can identify alternative splicing isoforms, gene fusions, single nucleotide variants, non-coding RNAs, and other transcriptomic features, enabling comprehensive exploration of the transcriptome.
- Qualitative and quantitative data: RNA-Seq generates both qualitative and quantitative data. It provides information about the presence or absence of transcripts (qualitative) as well as their relative abundance or expression levels (quantitative). This dual nature of RNA-Seq data enables researchers to gain insights into both the presence of different transcripts and their expression patterns.
- Complete transcriptome coverage: Unlike traditional methods that focus on a limited number of transcripts or predefined targets, RNA-Seq provides a comprehensive view of the entire transcriptome. It captures the expression profiles of all transcribed genes, including rare or previously uncharacterized transcripts. This unbiased approach allows for a more thorough understanding of gene regulation and transcriptomic changes.
- Species-independent applicability: RNA-Seq can be applied to any species, regardless of whether a reference sequence is available. It is particularly useful for studying non-model organisms or species with limited genomic resources. RNA-Seq enables de novo transcriptome assembly, where the transcriptome is reconstructed without relying on a reference genome, facilitating the study of novel species or those with complex genomes.
RNA sequencing empowers transcriptomics
RNA sequencing (RNA-Seq) has revolutionized transcriptome research in numerous fields, enabling scientists to gain valuable insights into gene expression, regulation, and functional analysis. Here are some ways in which RNA-Seq is empowering transcriptomics research across different areas:
- Disease research: RNA-Seq has significantly contributed to understanding the molecular basis of diseases. By comparing gene expression profiles between healthy and diseased tissues or cell types, researchers can identify differentially expressed genes that may play key roles in disease development, progression, or response to treatment. RNA-Seq also enables the discovery of disease-specific transcriptomic signatures and biomarkers, aiding in disease diagnosis, prognosis, and personalized medicine.
- Drug response and pharmacogenomics: RNA-Seq allows researchers to study how cells or tissues respond to drug treatments. By examining gene expression changes upon drug administration, researchers can identify molecular pathways involved in drug response, mechanisms of drug resistance, and potential side effects. Additionally, RNA-Seq can be used to investigate inter-individual differences in drug response, known as pharmacogenomics, by correlating gene expression profiles with treatment outcomes, helping to guide personalized therapies.
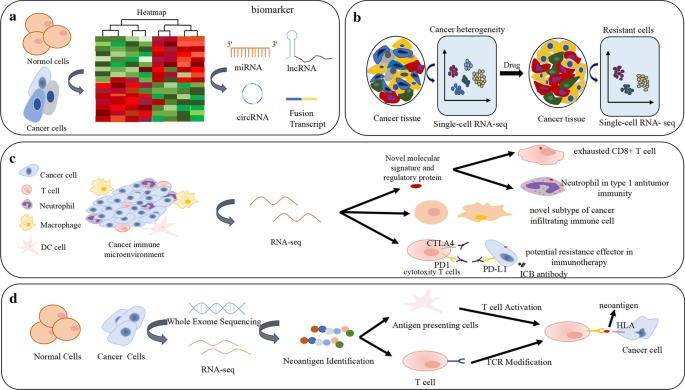 Applications of RNA-seq. (Hong et al., 2020)
Applications of RNA-seq. (Hong et al., 2020)
- Developmental biology: RNA-Seq is instrumental in unraveling the gene expression dynamics during various stages of development. By comparing transcriptomes across different developmental time points, researchers can identify genes and pathways involved in embryonic development, tissue differentiation, and organ formation. RNA-Seq can also reveal spatiotemporal expression patterns and regulatory networks, providing insights into the underlying mechanisms of development.
- Environmental responses: RNA-Seq allows researchers to study how organisms respond to different environmental conditions, such as temperature changes, exposure to pollutants, or nutrient availability. By analyzing gene expression patterns, we can identify genes and pathways involved in stress responses, adaptation, and environmental signaling. This information helps in understanding the molecular mechanisms of environmental responses and can have implications for fields like ecology, agriculture, and conservation biology.
- Gene regulation studies: RNA-Seq provides a comprehensive view of the transcriptome, enabling the study of gene regulation at different levels. It allows the identification of alternative splicing events, detection of non-coding RNAs, and investigation of post-transcriptional regulation. By integrating RNA-Seq data with other epigenomic and proteomic datasets, researchers can gain a more holistic understanding of gene regulation networks and the functional consequences of regulatory events.
In combination with other omics approaches, such as genomics, epigenomics, and proteomics, RNA-Seq provides a powerful tool to unravel the complexities of gene expression and regulation. It enables the identification of functional elements, novel transcripts, regulatory networks, and disease-associated pathways, ultimately contributing to our understanding of biological processes and human health.
Applications of RNA-Seq: learn how RNA sequencing is advancing transcriptome research in various fields
How to apply RNA sequencing methods?
RNA-Seq methods offer different advantages and are applicable to various research questions and experimental setups. Researchers can choose the most suitable method based on their specific objectives and available resources.
- mRNA Sequencing: This method allows for sensitive and accurate quantification of gene expression. It can identify known and novel isoforms in the coding transcriptome, detect gene fusions, and measure allele-specific expression.
- Targeted RNA Sequencing: This approach focuses on analyzing gene expression in a specific set of genes of interest. Targeted RNA-Seq can be achieved through enrichment methods, where RNA fragments corresponding to the target genes are enriched before sequencing, or through amplicon-based approaches, where specific regions of interest are amplified and sequenced.
- Ultra-Low-Input and Single-Cell RNA-Seq: This method is used to examine the signals and behavior of individual cells within their surrounding environment. It is particularly useful for studying processes such as cell differentiation, proliferation, and tumorigenesis. Ultra-low-input RNA-Seq enables analysis of very small amounts of starting material, while single-cell RNA-Seq allows for the characterization of gene expression in individual cells.
Single-Cell RNA Sequencing Q&A: learn more knowledge of Single-Cell RNA Sequencing.
Single-cell RNA Sequencing: Introduction, Methods, and Applications: see how to do your next scRNA-Seq experiment. - Total RNA Sequencing: This method provides a comprehensive view of the transcriptome by accurately measuring gene and transcript abundance. It can detect both known and novel features in coding RNA as well as various forms of noncoding RNA. More details, please refer to Total RNA Sequencing Q&A.
- Small RNA Sequencing: This technique focuses on isolating and sequencing small RNA species, such as microRNAs. It helps in understanding the role of noncoding RNA in gene silencing and posttranscriptional regulation of gene expression.
- LncRNA Sequencing: To explore the functions and characteristics of lncRNAs, we employ lncRNA sequencing coupled with high-throughput sequencing technology and bioinformatics analysis.
How to analyze RNA using NGS?
- Extract RNA from the sample.
- Prepare an RNA library for sequencing.
- Perform NGS to generate sequencing reads.
- Bioinformatics Workflow of RNA-Seq.
- Conducting quality control checks.
- Aligning reads to a reference genome/transcriptome.
- Quantifying gene/transcript expression levels.
- Performing differential expression analysis.
- Conducting functional analysis.
- Interpret and visualize the results.
Please refer to The Technologies and Workflow of RNA-Seq to help you determine the process most appropriate for your research.
Reference:
- Hong, Mingye, et al. "RNA sequencing: new technologies and applications in cancer research." Journal of hematology & oncology 13.1 (2020): 1-16.


 Sample Submission Guidelines
Sample Submission Guidelines
