Applications of Exome Sequencing in Plants
Exome sequencing, which zeroes in on the protein-coding portions of the genome, has emerged as a crucial technique in plant biology and agricultural advancement. By focusing on exons-regions that make up only 1–2% of the genome but contain nearly 85% of variants linked to diseases-scientists can effectively detect genetic differences that impact essential plant traits. This method is especially advantageous for species with large and complex genomes, where conducting whole-genome sequencing may prove less feasible due to higher costs and technical challenges.
Summary of Exome Sequencing
Exome sequencing is a powerful genomic technique that focuses on sequencing the protein-coding regions of an organism's genome. Unlike whole-genome sequencing (WGS), which examines the entire DNA sequence, exome sequencing specifically targets exons, the regions that encode functional proteins. Although exons make up only about 1–2% of the genome, they contain approximately 85% of disease-related and functionally significant genetic variations. This method provides a cost-effective and efficient approach to identifying genetic variations associated with key biological traits, particularly in plants with large and complex genomes.
In plant research, exome sequencing has revolutionized genetic studies by enabling the discovery of single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) that affect phenotypic characteristics. Researchers can efficiently link genetic variation to specific functions by focusing on coding regions, making exome sequencing a valuable tool in plant breeding, stress adaptation studies, and evolutionary research. Moreover, its application extends to identifying genes responsible for important agronomic traits, such as disease resistance, drought tolerance, and crop yield improvement. With next-generation sequencing technologies advancements, exome sequencing plays a crucial role in understanding plant genetics and improving agricultural sustainability.
Technical Principles of Exome Sequencing in Plants
Exome sequencing relies on targeted enrichment of exonic regions. Two primary strategies are used:
(1) Hybridization-Based Capture
Custom RNA or DNA probes (e.g., Agilent SureSelect, NimbleGen SeqCap) hybridize to exonic DNA fragments. Probes are designed using reference genomes or transcriptome data. For polyploid crops like wheat, this requires careful optimization to avoid cross-hybridization with homologous regions.
(2) PCR-Based Enrichment
Multiplex PCR amplifies target exons directly.
Ideal for small gene panels (e.g., screening 100 drought-related genes across 500 maize lines).
Modern exome sequencing predominantly employs short-read platforms like Illumina NovaSeq (2×150 bp reads) for high accuracy and affordability. A typical run generates ~5 GB of data per sample at 50–100× coverage-sufficient for reliable variant detection-compared to the 150 GB produced by WGS. Long-read technologies (PacBio HiFi, Oxford Nanopore) are increasingly integrated to resolve challenges such as splice variants. For example, combining Illumina exome data with Nanopore reads improved splice junction detection in soybean by 30%.
The bioinformatics pipeline begins with read alignment using tools like BWA-MEM, which maps sequences to a reference genome. Polyploid species, such as octoploid strawberries, demand specialized references to distinguish between homoeologs. Next, variant calling with GATK or SAMtools identifies SNPs and indels, followed by stringent filtering to exclude artifacts in repetitive regions. Functional annotation via databases like PlantCyc links variants to biological processes-for instance, a promoter SNP in the GmSWEET10a gene disrupts transcription factor binding, reducing soybean oil synthesis.
While exome sequencing offers unmatched cost efficiency and faster turnaround, it has limitations. It cannot detect structural variants (e.g., large deletions) or alternative splicing events, which account for 25% of protein diversity in tomatoes. To address this, hybrid approaches combining exome data with low-coverage WGS or RNA-seq are gaining traction, enabling comprehensive analyses at reduced costs. For orphan crops lacking high-quality genomes, transcriptome-guided probe design enhances capture accuracy, as shown in finger millet.
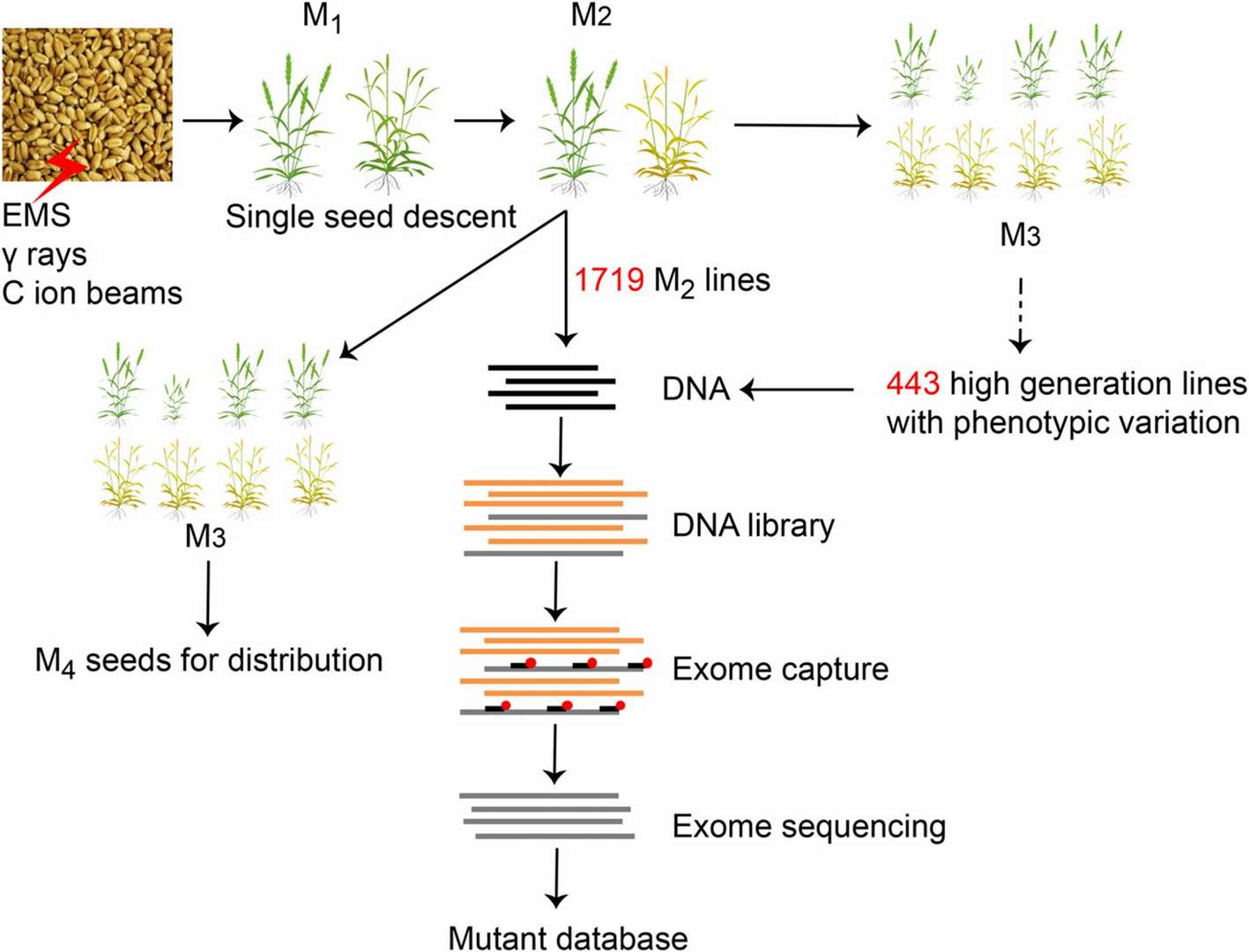 Figure 1.Overview of the method used to establish the wheat mutant database by exome sequencing.(Hongchun Xiong .2023)
Figure 1.Overview of the method used to establish the wheat mutant database by exome sequencing.(Hongchun Xiong .2023)
Services you may interested in
Learn More
Applications of Exome Sequencing in Plant Research
Exome sequencing has emerged as a transformative tool in plant genetic research, significantly enhancing our ability to identify genetic variations that underpin key agronomic traits. One of the most impactful applications of this technology is in mutation detection for plant breeding. A notable example is the study on Nicotiana tabacum L. (tobacco), where WES was employed to identify ethyl methanesulfonate (EMS)-induced mutations. This approach not only provided a cost-effective alternative to WGS but also demonstrated the potential of exome sequencing in high-throughput mutation identification.
Case Study: Mutation Detection in Tobacco
Researchers applied WES to investigate EMS-induced mutations in 19 independent M2 tobacco lines, aiming to develop a high-throughput mutation identification pipeline while optimizing detection efficiency. Tobacco, being a complex allotetraploid species with a large genome size of 4.5 Gb, posed significant challenges for whole-genome sequencing. Exome sequencing emerged as a strategic choice, allowing researchers to focus on protein-coding regions and significantly reduce sequencing costs. Through this approach, they were able to identify numerous SNPs and indels within coding regions, which are often associated with phenotypic variations. This study not only highlighted the utility of exome sequencing in mutation detection but also demonstrated its potential for improving breeding efficiency.
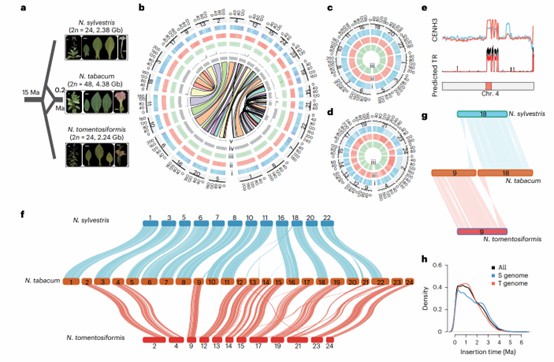 Figure 2. Comparative genomic analysis by exome sequencing.(Zan, Y., 2025)
Figure 2. Comparative genomic analysis by exome sequencing.(Zan, Y., 2025)
Applications in Plant Breeding
Exome sequencing has revolutionized plant breeding by enabling the identification of alleles associated with desirable traits. In rice, for instance, exome sequencing has been used to detect genes related to yield and disease resistance. By focusing on protein-coding regions, researchers can efficiently link genetic variations to specific functions, facilitating the development of improved cultivars through marker-assisted selection. This approach has been particularly valuable in crops with large and complex genomes, where traditional breeding methods may be less efficient.
Adaptation and Resistance Gene Discovery
Understanding how plants adapt to environmental stresses is crucial for developing resilient crops. Exome sequencing has been employed to identify genes responsible for abiotic stress tolerance in crops like barley, shedding light on mechanisms of drought and salinity resistance. For example, researchers have used exome sequencing to uncover genetic variations linked to drought tolerance in pigeonpea. These findings provide valuable markers for selective breeding, enabling the development of crop varieties that can better withstand adverse environmental conditions.
Crop Domestication and Evolutionary Studies
Exome sequencing also provides insights into the domestication processes of crops by comparing wild and cultivated varieties. Research on maize has revealed genetic changes associated with domestication traits, enhancing our understanding of crop evolution. By focusing on protein-coding regions, exome sequencing allows researchers to identify genes that have been selected during domestication, providing valuable information for breeding programs and conservation efforts.
Development and Utilization of Plant Resources
In addition to its applications in crop improvement and stress adaptation studies, exome sequencing has also been instrumental in developing and utilizing plant resources. In medicinal plants, for example, exome sequencing facilitates the discovery of genes involved in the biosynthesis of therapeutic compounds. A notable example is Artemisia annua, the plant source of artemisinin, a potent antimalarial compound. Exome sequencing has identified key enzymes in the artemisinin biosynthetic pathway, aiding in the development of high-yielding varieties. This application enhances the production of valuable medicinal compounds and contributes to the sustainable use of plant resources.
Comparison of Whole Genome Sequencing and Exome Sequencing
Exome sequencing, which targets only the exons of the genome, offers several key advantages over WGS:
1) Cost-Effectiveness – By sequencing only ~1-2% of the genome, exome sequencing significantly reduces costs while still capturing most disease- or trait-relevant variants.
2) Higher Depth & Accuracy – Exome sequencing achieves much higher read depth with the same sequencing effort, improving variant detection accuracy, especially for rare mutations.
3) Simpler Data Analysis – The smaller dataset size reduces computational burden and simplifies bioinformatics processing compared to WGS.
4) Better for Mendelian & Functional Studies – Since most known disease-causing mutations occur in exons, exome sequencing is highly efficient for identifying pathogenic variants in genetic disorders and agronomic traits.
5) Proven Utility in Plants – Exome sequencing has been successfully applied in crops (e.g., wheat, rice) to identify genes linked to yield, stress resistance, and quality traits without the complexity of whole-genome analysis.
While WGS provides comprehensive genomic data, exome sequencing remains a powerful, targeted approach for functional genomics and precision breeding.
This study presents a comprehensive genomic analysis of 472 Vitis accessions, spanning 48 out of 60 extant grapevine species, using whole-genome resequencing to decode genetic variation at single-base resolution. It reveals dynamic population histories, including a dramatic expansion and contraction in domesticated grapevines and unique demographic patterns in pan-Black Sea cultivars. Key selective sweeps were identified for traits like berry edibility and stress resistance, alongside gene-trait associations for berry shape and aromatic compounds, offering critical insights for grapevine breeding. While this work employed whole-genome sequencing, exome sequencing-a cost-effective alternative targeting protein-coding regions-is widely used in plant studies to efficiently identify functional variants linked to agronomic traits, such as disease resistance or yield optimization, particularly in species with large or complex genomes. Both approaches accelerate the discovery of evolutionary and breeding-relevant genetic signatures.
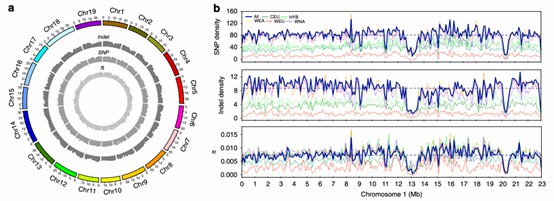 Figure 3. Summary of the genomic variations identified across 472 Vitis accessions.( Liang, Z,et. al,2019)
Figure 3. Summary of the genomic variations identified across 472 Vitis accessions.( Liang, Z,et. al,2019)
Advantages of Exome Sequencing in Plant Research
Exome sequencing offers several advantages for plant researchers. Firstly, its targeted approach reduces the amount of sequencing data needed compared to whole-genome sequencing, making it more cost-effective and less computationally intensive. This is especially beneficial for plants with large genomes, such as wheat or maize, where whole-genome sequencing can be prohibitively expensive and complex. Secondly, exome sequencing allows researchers to identify functional genetic variations more efficiently by focusing on exons. This is crucial for understanding the genetic basis of important traits, such as disease resistance, stress tolerance, and yield improvement.
Challenges and Future Directions
While exome sequencing has significantly advanced plant research, it also faces several challenges. One of the main challenges is the incomplete capture of exonic regions, which can lead to missing important genetic variations. Additionally, the interpretation of genetic variations identified through exome sequencing can be complex, especially when the function of a gene is not well understood. Future advancements in sequencing technologies and bioinformatics tools will be crucial in addressing these challenges. For example, improvements in capture efficiency and the development of more accurate variant calling algorithms will enhance the effectiveness of exome sequencing. Furthermore, integrating exome sequencing data with other omics data, such as transcriptomics and proteomics, will provide a more comprehensive understanding of plant genetics and biology.
In conclusion, exome sequencing has become an indispensable tool in plant research, offering a powerful approach to identifying genetic variations associated with key biological traits. Its applications in plant breeding, stress adaptation studies, crop domestication, and the development of plant resources have significantly advanced our understanding of plant genetics. As sequencing technologies continue to improve, exome sequencing will undoubtedly play an even more important role in improving agricultural sustainability and addressing global food security challenges.
Whole exome sequencing offers numerous advantages, but challenges remain:
- Data Analysis: Managing and interpreting large datasets requires advanced bioinformatics tools and expertise.
- Incomplete Annotations: In some plants, incomplete genome annotations can hinder accurate exon capture and variant identification.
- Integration with Other Omics Data: Combining exome sequencing data with transcriptomics and epigenomics is essential for a comprehensive understanding of gene regulation but adds complexity to data analysis.
Conclusion
Exome sequencing has become an indispensable tool in plant genomics, offering detailed insights into the coding regions that govern vital traits. Its applications in understanding genetic variation, aiding plant breeding, uncovering adaptation mechanisms, studying domestication, and exploring valuable plant resources underscore its significance. As technological advancements continue to reduce costs and improve data analysis, the integration of exome sequencing into plant research is poised to expand, contributing to sustainable agriculture and food security. Exome sequencing has revolutionized plant research by providing a powerful tool to identify genetic variations associated with key biological traits. Its applications in plant breeding, stress adaptation studies, and evolutionary research have significantly advanced our understanding of plant genetics. As sequencing technologies continue to improve, exome sequencing will undoubtedly play an even more important role in improving agricultural sustainability and addressing global food security challenges.
References:
- Xiong, H., Guo, H., et,al., A large-scale whole-exome sequencing mutant resource for functional genomics in wheat. Plant biotechnology journal, 2023; 21(10), 2047–2056. https://doi.org/10.1111/pbi.14111
- Zan, Y., Chen, S., et al. The genome and GeneBank genomics of allotetraploid Nicotiana tabacum provide insights into genome evolution and complex trait regulation. Nat Genet (2025). https://doi.org/10.1038/s41588-025-02126-0
- Liang, Z., Duan, S., et,al., (2019). Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nature communications, 10(1), 1190. https://doi.org/10.1038/s41467-019-09135-8


 Sample Submission Guidelines
Sample Submission Guidelines
