Evolutionary, Medical, and Clinical Applications of Whole Exome Sequencing
As we all know, proteins play important roles in human bodies since all the physiologic events are related to the structural or mechanical functions of certain proteins. To study the synthesis process of proteins is a way to study the etiology, pathophysiology, diagnosis and treatment of certain diseases. The human genome contains approximately 3 billion base pairs of DNAs. Among them, only about 30 million base pairs of DNAs are carrying the information of protein synthesis. This part of genome is called exome. Exome is valuable in researchers' eyes as it is the origin of protein synthesis. With the development of exome-enrichment strategies and high-throughput sequencing, whole exome sequencing (WES) becomes possible. WES is a more cost-effective method compared to whole genome sequencing, and has been applied in evolutionary and medical research, and clinical practice.
Application in evolution research
Genome is an encyclopedia, telling us the stories happened throughout the history of human beings. Whole exome sequencing could be helpful by solving the puzzle of human evolution. By studying anthropology, intervention of some diseases could be more efficient. Take esophageal carcinoma as example. In China, people from two areas have a high incidence of esophageal carcinoma, around Tai Mountain and Chaoshan area in Guangdong Province. The languages, customs, lifestyles, foods and climates are quite different from each other. Historical study suggests that they may share the same ancients. Whole exome sequencing has been applied so as to figure out a gene expressing map of people from these two areas.
Application in medical research
Etiology research. There are lots of untreated diseases with unknown etiology which are called idiopathic diseases in clinic. Without knowing the causes, these diseases could only be diagnosed and treated according to symptoms, such as primary hypertension and primary amenorrhea. All we know, these diseases are not secondary. Doctors used to study these diseases by vital sign monitoring and laboratory examination result of patients. Then with the development of biological technology, the physiological alternation is found at tissue, cellular or even molecular levels. Lots of idiopathic diseases are proved to be related to dysfunction or deficiency of certain protein. For further analysis of the etiology of idiopathic diseases, whole exome sequencing allows us to figure out the origin of protein dysfunction or deficiency.
One example is lysosomal storage diseases. This group of diseases is defined as the deficient function of lysosomal resulting in delay growth, deafness or dementia. Lysosomal is a kind of organelle which breaks down certain substances inside cells. We know little about the mechanism of lysosomal storage diseases. Nowadays, researchers have found several genes related to this group of disease by using whole exome sequencing. By knowing the functioning genes, exploration like gene knock-out experiments can be performed to further understand the mechanism of this group of disease. Another example is cancer with unknown etiology. By analyzing the etiology of cancer, doctors could figure out the prevention strategies. Figure 1 shows a typical etiology research using whole exome sequencing.
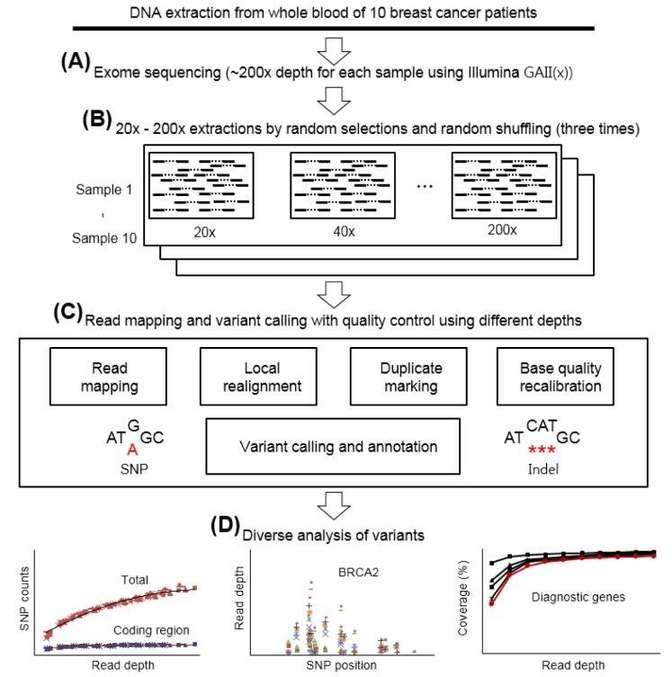
Figure 1. A typical research of breast cancer using whole exome sequencing. (Kim et al, 2015)
Epidemiology research. Risks of getting certain diseases vary among individuals. Some of the diseases are easily developed in certain groups of people. The variety of incidence is due to the differences in sanitary condition, culture and genetic factors. It is easy to compare the sanitary condition and culture by social study. But it is difficult to study the genetic factors as the quantity of genome is too large. Current psychiatrists benefit a lot from whole exome sequencing, as it solves the problem above.
Take the Alzheimer's disease as an example. Alzheimer's disease is a chronic degenerative disease resulting in dementia. Symptoms like memory loss usually happen to old people. When they get older, the symptoms become severer. It is an irreversible progress. What we can do is early intervention. However, people could hardly distinguish whether their grandfather has behavior change or just in a bad mood, unless those who are trained. This prevents the early intervention. There is a project call Alzheimer's Disease Sequencing Project, which would perform whole exome sequencing in more than 5 thousand of patients, so as to draw out genes responsible for Alzheimer's diseases. This will be a strong supportive material in epidemiologic practice, as the local government could come up with preventing project more specifically according to the gene expressing condition of the residents.

Figure 2. Alzheimer’s Disease Sequencing Project.
Application in clinical practice
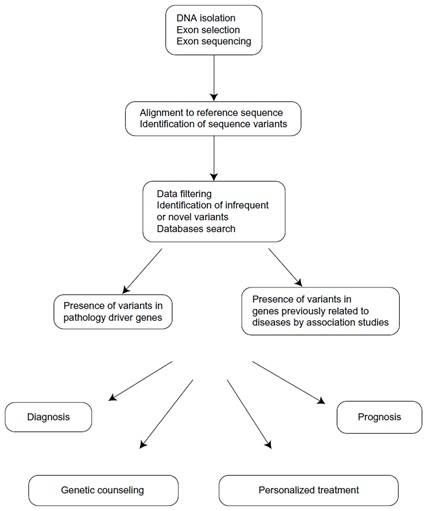
Figure 3. Applications of whole exome sequencing in clinic.
Diagnosis. Diagnosis is the most important process in clinical practice. It requires doctors to be familiar with symptoms, signs, laboratory indicators and imageological changes of certain diseases. Sometimes the patient will misunderstand doctors' questions, resulting in providing a confusing history to doctors. This may happen when the paitient is low-educated, delirious, or infant, probably leading to misdiagnosis. Whole exome sequencing makes great contributions in these cases. Doctors could use whole exome sequencing to diagnose and to avoid misdiagnosis. There are evidences that whole exome sequencing has been applied to pediatric department, helping to diagnose neurologic diseases in newborns and infants. Also, whole exome sequencing is used to prognose and diagnose and mosaic variegated aneuploidy syndrome by detecting CEP57 mutation.
Treatment. Treatment to a certain disease does not always work, especially for those with systemic diseases like hypertension, diabetes and cancers. Take cancers as an example. At the first time, we know cancer results from uncontrolled growth of cells, but we do not know which specific type of cell is. So we use chemotherapy, radiotherapy or surgery to eliminate the abnormal tissue which we call nidus. These treatments may not cure the patients sometimes, especially in malignant cases. Also, all these treatments could kill normal tissue as well as the abnormal ones, which may worsen the condition of patients. With the development of clinical and basic medicine, the understanding of the pathophysiology of carcinoma gets deeper. We know some potential target of cancers, such as EGFR, mTOR and KEAP1. Targeted therapy aimed at these loci strikes the abnormal cells precisely, and spares the normal cells. It maximizes the effect of treatment and minimizes the side effect. By whole exome sequencing, we could know the classification of tumor so as to come up with a personalized targeted therapy.
Prevention. It is more effective to prevent than to treat. However, not all the diseases show a clear early symptom. Whole exome sequencing provides the gene profile of a person. With this gene profile, one could know his or her risk factors of certain diseases so he or she could prevent it early. For example, female knowing that she has mutations in DYNC2H1 and ALOX15, which is known to be a risk factor of recurrent early pregnancy loss, may turn to her obstetrician before she will plan to have a baby.
Additional reading:
Principles and Workflow of Whole Exome Sequencing
Bioinformatics Workflow of Whole Exome Sequencing
References:
- Kim K., Seong M., Chung W., Park S.S., Leem S., Park W., Kim J, Lee, K., Park R.W. and Kim N. Effect of Next-Generation Exome Sequencing Depth for Discovery of Diagnostic Variant. Genomics Inform. 2015 Jun; 13(2): 31–39.
- Tang W.R., Chen Z.J., Lin K., Su M. and Au W.W. Development of esophageal cancer in Chaoshan region, China: association with environmental, genetic and cultural factors. Int J Hyg Environ Health. 2015 Jan;218(1):12-8.
- Nolan D and Carlson M. Whole Exome Sequencing in Pediatric Neurology Patients: Clinical Implications and Estimated Cost Analysis. J Child Neurol.2016 Jun;31(7):887-94.
- Qiao Y, Wen J, Tang F, Martell S, Shomer N and Leung P.C., Stephenson M.D., Rajcan-Separovic E. Whole exome sequencing in recurrent early pregnancy loss. Mol Hum Reprod. 2016 May;22(5):364-72.
- Tian T, Lu Y, Yao J, Cao X, Wei Q and Li Q. Identification of a novel MYO6 mutation associated with autosomal dominant non-syndromic hearing loss in a Chinese family by whole-exome sequencing. Genes Genet Syst. 2018 Aug 31.
- Lu T, Li M, Xu X, Xiong J, Huang C, Zhang X, Hu A, Peng L, Cai D, Zhang L, Wu B and Xiong F. Whole exome sequencing identifies an AMBN missense mutation causing severe autosomal-dominant amelogenesis imperfecta and dentin disorders.Int J Oral Sci. 2018 Sep 3;10(3):26.
- Klapproth E, Dickreuter E, Zakrzewski F, Seifert M, Petzold A, Dahl A, Schröck E, Klink B, Cordes N.Whole exome sequencing identifies mTOR and KEAP1 as potential targets for radiosensitization of HNSCC cells refractory to EGFR and β1 integrin inhibition. Oncotarget. 2018 Apr 6;9(26):18099-18114.
- Wang N, Zhang Y, Gedvilaite E, Loh JW, Lin T, Liu X, Liu CG, Kumar D, Donnelly R, Raymond K, Schuchman EH, Sleat DE, Lobel P, Xing J. Using whole-exome sequencing to investigate the genetic bases of lysosomal storage diseases of unknown etiology. Hum Mutat. 2017 Nov;38(11):1491-1499.
- Zhu Q, Hu Q, Shepherd L, Wang J, Wei L, Morrison CD, Conroy JM, Glenn ST, Davis W, Kwan ML, Ergas IJ, Roh JM, Kushi LH, Ambrosone CB, Liu S, Yao S. The impact of DNA input amount and DNA source on the performance of whole-exome sequencing in cancer epidemiology. Cancer Epidemiol Biomarkers Prev. 2015 Aug;24(8):1207-13.
- Sánchez-Quinto F and Lalueza-Fox C. Almost 20 years of Neanderthal palaeogenetics: adaptation, admixture, diversity, demography and extinction. Philos Trans R Soc Lond B Biol Sci. 2015 Jan 19;370(1660):20130374.


 Sample Submission Guidelines
Sample Submission Guidelines
