Microbiome research is transitioning from a "species census" to an era of "functional decoding," where the choice of technology directly determines the depth and boundaries of scientific inquiry. Metagenomic shotgun sequencing and metatranscriptomic sequencing serve as complementary counterparts, akin to a "static map" of the genome and a "dynamic image" of the transcriptome: the former randomly captures microbial DNA fragments to reveal the species composition and functional potential of a community, while the latter focuses on the total RNA (primarily mRNA) within environmental samples to analyze real-time gene expression and metabolic activity of microorganisms. These two techniques, from the perspectives of genomic potential and transcriptomic activity, together construct the "double helix" of microbiome research. However, differences in experimental design, data interpretation, and application scenarios often lead researchers to face the dilemma of choosing between DNA and RNA. This article aims to provide a precise guide for functional microbiome research through a detailed comparison of the technical principles, application contexts, and case studies of both methods.
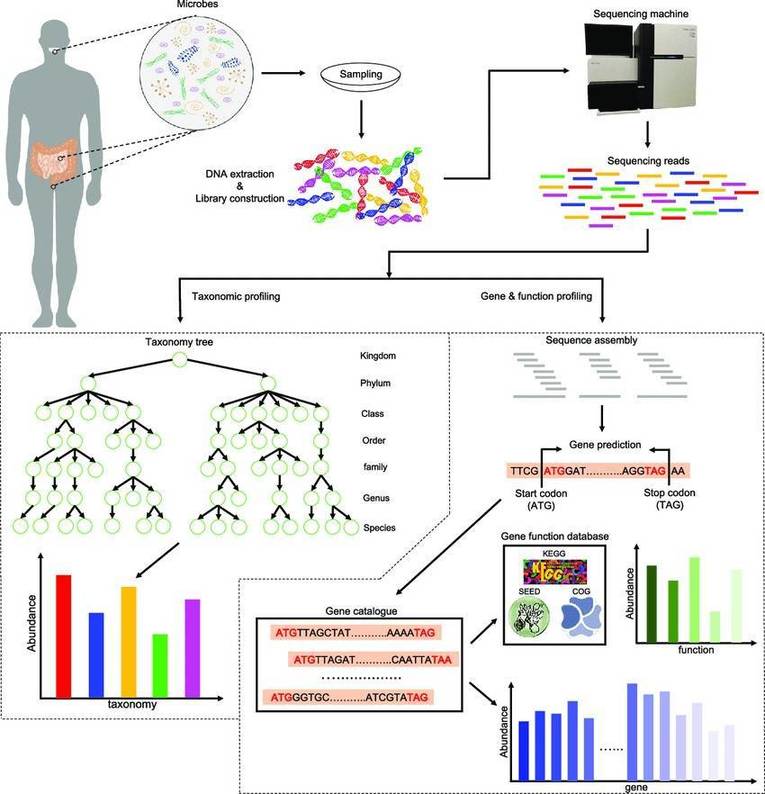
 Effects of the gut microbiome on diseases and international metagenomics and metatranscriptomics consortia and projects.
Effects of the gut microbiome on diseases and international metagenomics and metatranscriptomics consortia and projects.
Metagenomic Shotgun Sequencing
Principle and Experimental Design
Metagenomic shotgun sequencing involves the random fragmentation of total DNA from environmental samples (including bacteria, archaea, fungi, viruses, etc.), followed by high-throughput sequencing for unbiased fragment capture. The data is then analyzed using bioinformatics tools to reconstruct the microbial community's species composition and functional gene repertoire.
Key Features:
- DNA-Level Analysis: Reveals the genetic potential of microbial communities, answering the question, "What can they do?"
- No Amplification Bias: PCR is not required, minimizing biases related to species abundance during sequencing.
- Functional Gene Annotation: Uses databases such as KEGG and COG to predict metabolic pathways (e.g., antibiotic resistance genes, carbon and nitrogen cycle genes).
Application Scenarios:
- Comprehensive Survey of Species and Functional Genes: For example, functional potential analysis of gut microbiota.
- Discovery of Novel Species or Pathogens: Such as genome mining of microorganisms from deep-sea or extreme environments.
- Tracking Antibiotic Resistance Genes (ARGs) or Biosynthetic Gene Clusters (BGCs): For investigating resistance mechanisms or novel bioactive compound production.
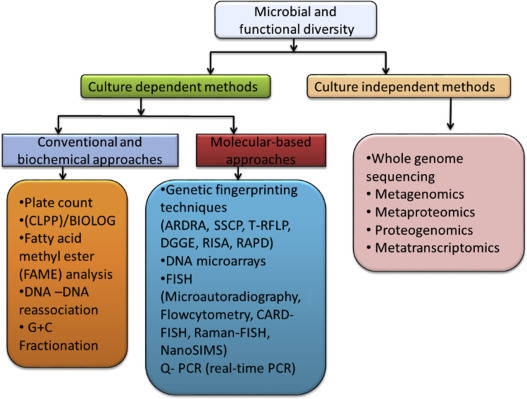
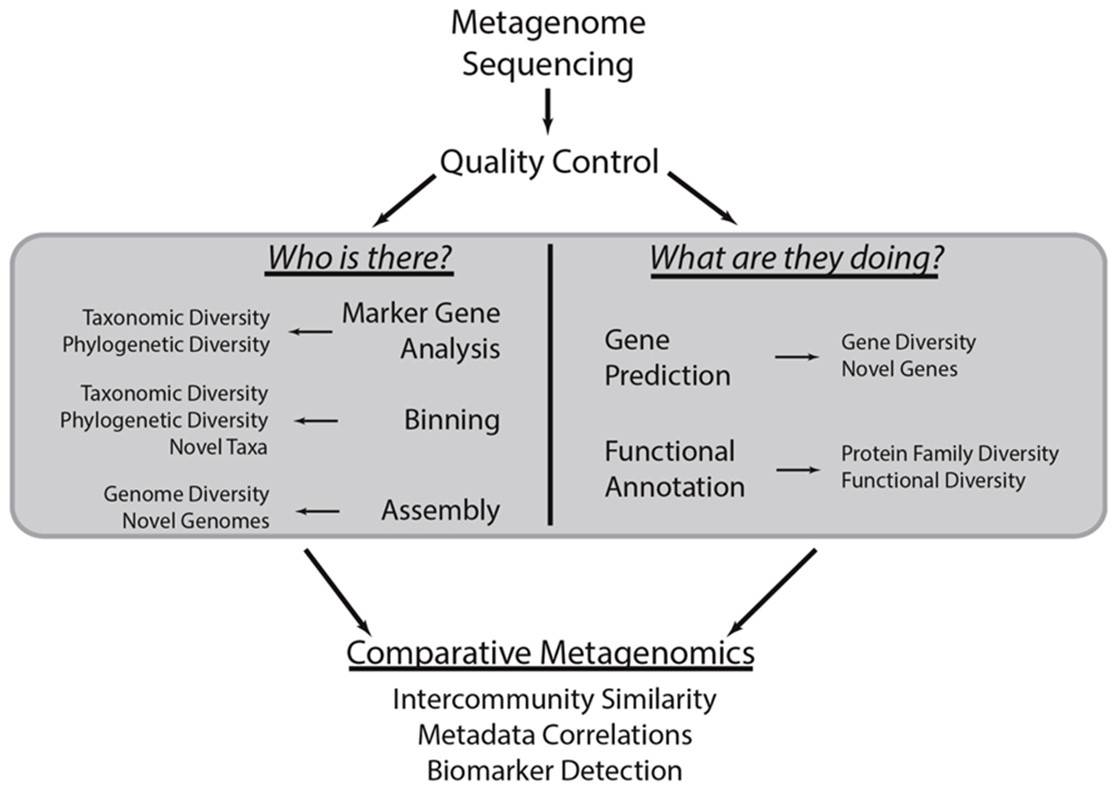 Common metagenomic analytical strategies.
Common metagenomic analytical strategies.
Case Studies:
| Research Topic | Journal & Year | Key Findings & Applications | Technical Highlights |
|---|---|---|---|
| Revolutionary Discovery of Marine Microbial Diversity | Science (2004) | Conducted large-scale shotgun sequencing of the Sargasso Sea microbiome, identifying over 1,800 genomic species (including 148 novel bacterial clades) and more than 1.2 million unknown genes, revealing complex metabolic networks (e.g., novel photoreceptor genes) in marine microbes. | First unbiased coverage of marine microbial genomes, establishing a standard analysis framework for environmental metagenomics. |
| Cross-Environmental Microbial Functional Comparison | Science (2004) | By comparing metagenomes from soils, oceans, and other environments, identified environment-specific functional gene signatures (e.g., land enriched in lignin degradation genes, oceans enriched in sulfur metabolism genes), demonstrating the role of horizontal gene transfer in environmental adaptation. | Developed gene-centric comparative analysis methods, advancing ecological function predictions. |
| Global Microbiome Atlas Construction | Nature Biotechnology (2020) | Integrated shotgun data from over 10,000 environmental samples to publish the Global Earth Microbiome Catalog (GEM Catalog), encompassing over 15,000 high-quality metagenome-assembled genomes (MAGs), expanding the tree of life. | Used binning techniques to extract single microbial genomes from complex samples. |
| Human Microbiome Functional Analysis | Nature (2012) | As part of the Human Microbiome Project, shotgun sequencing revealed the association between short-chain fatty acid synthesis gene clusters in the gut microbiota and host immune regulation, advancing personalized probiotics development. | Conducted comparative metagenomics across body sites to develop host-microbe co-evolution models. |
Services you may interested in
Learn more:
Metatranscriptomic Sequencing
Principles and Experimental Design
Metatranscriptomic sequencing involves the extraction of total RNA from environmental samples, with a focus on enriching mRNA while removing ribosomal RNA (rRNA). The resulting RNA is then used for library construction and sequencing, enabling the analysis of gene expression activity within microbial communities.
Core Characteristics
- RNA-Level Analysis: Provides insights into the real-time metabolic state of microorganisms, answering the question of what they are actively doing.
- High Dynamic Sensitivity: Capable of detecting the impact of environmental disturbances (e.g., temperature shifts, host immune responses) on gene expression.
- Functional Validation Tool: Complements shotgun metagenomics by confirming whether specific functional genes are activated, such as monitoring the expression of virulence factors.
Application Scenarios
- Microbial Community Activity and Metabolic Pathways: Analysis of gene expression differences within microbial communities under varying conditions (e.g., examining how gut microbiota's gene expression varies in disease states).
- Environmental Stress Response Studies: Investigating the effects of environmental stressors (e.g., ocean acidification) on gene expression related to key microbial functions (e.g., carbon fixation in marine plankton).
- Host-Microbe Interaction Mechanisms: Real-time monitoring of pathogen virulence gene expression to study host-pathogen dynamics.
Case Study
| Application Scenario | Case Description | Reference | DOI Link |
|---|---|---|---|
| Active Metabolic Pathway Analysis | A study by the Nature Microbiology research team performed metatranscriptomic analysis on fecal samples from 308 elderly male subjects. The study found that core metabolic pathways (e.g., glycolysis, amino acid synthesis) were highly conserved across individuals, while more variable pathways, such as terpenoid synthesis, were associated with host health status. This research revealed the molecular basis of gut microbiota functional stability. | Abu-Ali, G.S. et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nature Microbiology (2018). | 10.1038/s41564-017-0084-4 |
| Environmental Stress Response Studies | A study published in PNAS demonstrated that ocean acidification significantly inhibited the expression of carbon fixation genes, such as RuBisCO, in marine plankton, while promoting the transcription of nitrogen metabolism-related genes. This revealed the dynamic regulatory mechanisms of microbial communities in balancing the carbon-nitrogen cycle. | Doney, S.C. et al. Climate change impacts on marine ecosystems. PNAS (2012). | https://doi.org/10.1146/annurev-marine-041911-111611 |
| Host-Microbe Interaction Mechanisms | Research published in Cell Host & Microbe using metatranscriptomics identified that host inflammatory signals, such as the IL-23/IL-17 axis, specifically activate virulence genes of Salmonella (e.g., SPI-1 and SPI-2 pathogenicity island genes), revealing the molecular mechanisms through which host immunity drives pathogen infection. | Thiennimitr, P. et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Cell Host & Microbe (2011). | 10.1073/pnas.1107857108 |
Overview of Comparison:
| Feature | Metagenomic Shotgun Sequencing | Metatranscriptomics |
|---|---|---|
| Target Molecule | DNA (Genomes) | RNA (Transcriptome, predominantly mRNA) |
| Information Type | Species composition, functional gene potential | Gene expression activity, real-time metabolic status |
| Technical Challenges | Host DNA interference (e.g., host >90% DNA in sample needs host removal) | RNA stability issues, rRNA removal efficiency affecting data quality |
| Functional Annotation Capability | Predicts potential metabolic pathways (based on gene presence) | Reveals active metabolic pathways (based on gene expression) |
| Time Resolution | Static (reflects the genomic state at the time of sampling) | Dynamic (reflects transcriptomic activity at the time of sampling) |
| Typical Cost (Per Sample) | $500–1500 (varies with sequencing depth) | $800–2000 (including RNA extraction and library preparation complexity) |
Differences in Technical Workflow
| Step | Metagenomic Shotgun Sequencing | Metatranscriptomic Sequencing |
|---|---|---|
| Starting Material | DNA | RNA (requires stable extraction and rRNA removal) |
| Fragmentation | Random fragmentation via sonication or enzymatic digestion | mRNA fragmentation or cDNA synthesis followed by fragmentation |
| Library Construction | Direct library construction from DNA fragments | Requires reverse transcription to cDNA prior to library construction |
| Key Challenges | Host DNA contamination, complex data assembly | RNA degradation, rRNA removal efficiency, host RNA contamination |
| Analysis Focus | Species composition, functional genes, antibiotic/virulence genes | Gene expression levels, active metabolic pathways, differentially expressed genes |
Application Scenarios Comparison
| Application Area | Metagenomic Shotgun Sequencing | Metatranscriptomic Sequencing |
|---|---|---|
| Clinical Diagnosis | Broad-spectrum pathogen detection (bacteria, viruses, fungi) and antibiotic resistance gene analysis | Investigating host-pathogen interactions, microbial activity under drug treatment |
| Environmental Microbiology | Species diversity, functional metabolic potential in environmental samples | Gene expression responses to environmental changes (e.g., temperature, pollution) |
| Basic Research | New species discovery, strain typing, genome assembly | Dynamics of transcriptional regulation, active pathway discovery |
| Biomarker Discovery | Stable biomarkers based on DNA (e.g., species abundance, functional genes) | Expression-based biomarkers (e.g., differentially expressed genes, metabolic activity) |
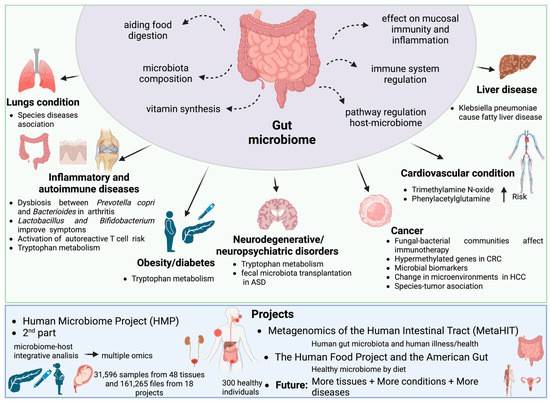
 The applications of metagenomics and metatranscriptomics in periodontitis: biomarker detection, treatment evaluation and connection with periodontitis-related systemic diseases.
The applications of metagenomics and metatranscriptomics in periodontitis: biomarker detection, treatment evaluation and connection with periodontitis-related systemic diseases.
(Yi Huang)
Differences in Data Volume and Analysis Methods
1. Data Volume
- Metagenomics: Typically generates large datasets due to comprehensive genome-wide coverage, requiring high sequencing depth to detect low-abundance species.
- Metatranscriptomics: Produces relatively smaller datasets but requires deeper sequencing to capture low-abundance transcripts with low expression levels.
2. Analysis Methods
Metagenomics:
- Reference-based classification: Tools such as MetaPhlAn2 and Kraken2 for species identification, and HUMAnN2 for functional annotation.
- Genome Assembly: Use of binning techniques to assemble microbial genomes from complex samples.
Metatranscriptomics:
- Expression Quantification: Calculation of transcript abundance (e.g., FPKM, TPM).
- Differential Analysis: Use of tests like Wilcoxon and LEfSe to identify differentially expressed genes.
- Functional Enrichment: Annotation of active pathways using KEGG and GO.
Advantages and Disadvantages Comparison
| Aspect | Metagenomic Shotgun Sequencing | Metatranscriptomic Sequencing |
|---|---|---|
| Advantages | 1. Unbiased detection of all microbial species and functional genes. | 1. Reflects real-time gene activity, distinguishing between active and inactive microbes. |
| 2. High resolution (down to strain level) and ability to discover novel species. | 2. Reveals dynamic transcriptional regulation mechanisms. | |
| 3. High data stability (DNA is more stable than RNA). | 3. Directly links phenotype to functional expression. | |
| Disadvantages | 1. Unable to distinguish microbial activity states. | 1. RNA is prone to degradation, requiring strict sample handling. |
| 2. Data analysis is complex and relies heavily on reference databases. | 2. High-expressed genes may obscure low-abundance transcripts. | |
| 3. Higher costs, particularly for large-scale samples. | 3. Requires effective rRNA and host RNA removal. |
Strategic Approach and Combined Application
1. Functional Potential vs. Activity Validation
- Functional Potential: When a comprehensive understanding of the microbial community's "functional toolkit" is required, metagenomic shotgun sequencing should be prioritized.
- Activity Validation: To determine the "current functional state" of microbes, metatranscriptomic sequencing is essential.
2. Multi-Omics Integration
- Shotgun Metagenomics + Transcriptomics: For instance, shotgun sequencing can be used to identify sulfur metabolism genes in hydrothermal vent communities, followed by transcriptomic analysis to assess the expression levels of these genes under high-temperature and high-pressure conditions.
- Shotgun Metagenomics + Metabolomics: This approach links the functional potential of microbial communities with their metabolite profiles (e.g., short-chain fatty acids), thereby providing evidence of functional execution.
3. Cost Optimization
For projects with budgetary constraints, shotgun metagenomics can initially be employed to identify key samples, followed by metatranscriptomic sequencing on a smaller subset of samples for deeper analysis.
Summary and Recommendations for Selection
- Metagenomic Shotgun Sequencing (mNGS): Recommended when comprehensive analysis of microbial species composition, functional potential, or discovery of unknown pathogens is needed, particularly in scenarios where DNA samples are stable and sufficient budget is available.
- Metatranscriptomic Sequencing: Suitable when the focus is on understanding the active functional status of microbes, environmental responses, or disease dynamics, provided that RNA quality is ensured and expertise in handling complex transcriptomic data is available.
- Combined Approach: Integrating both methods offers a comprehensive perspective on microbial communities, capturing both genetic potential and real-time functional activity. For example, in gut microbiota research, metagenomic shotgun sequencing provides species-level context, while metatranscriptomics uncovers metabolic activity.
References
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Comparative metagenomics of microbial communities. Science. 2005 Apr 22;308(5721):554-7. DOI: 10.1126/science.1107851. PMID: 15845853.
- Goma-Tchimbakala, E.J.C.D.; Pietrini, I.; Goma-Tchimbakala, J.; Corgnati, S.P. Use of Shotgun Metagenomics to Assess the Microbial Diversity and Hydrocarbons Degrading Functions of Auto-Mechanic Workshops Soils Polluted with Gasoline and Diesel Fuel. Microorganisms 2023, 11, 722. https://doi.org/10.3390/microorganisms11030722
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013 Jan;41(Database issue):D590-6. DOI: 10.1093/nar/gks1219. Epub 2012 Nov 28. PMID: 23193283; PMCID: PMC3531112.
- Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmento H, Hingamp P, Ogata H, de Vargas C, Lima-Mendez G, Raes J, Poulain J, Jaillon O, Wincker P, Kandels-Lewis S, Karsenti E, Bork P, Acinas SG. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol. 2014 Sep;16(9):2659-71. DOI: 10.1111/1462-2920.12250. Epub 2013 Sep 18. PMID: 24102695.
- Terrón-Camero, L.C.; Gordillo-González, F.; Salas-Espejo, E.; Andrés-León, E. Comparison of Metagenomics and Metatranscriptomics Tools: A Guide to Making the Right Choice. Genes 2022, 13, 2280. https://doi.org/10.3390/genes13122280