Operational taxonomic unit or OTU is considered as the basic unit used in numerical taxonomy. These units may refer to an individual, species, genus, or class. In numerical methods, the taxonomic units used are constantly not comparable to the formal taxonomic units. Therefore, the term operational taxonomic unit was coined. Since data is lacking on the correct OTUs in researches, various experiments have been conducted to evaluate the clustering method performance such as the rationality of OTU structure, the ability to cope with inflation of OTU, the stability of OTU, and the computational efficiency or runtime and memory requirements.
OTUs or clusters with sequence similarity in the molecular level are widely accepted as an analytical unit used in microbial ecology. All throughout the taxonomic development, OTUs are the most commonly used units when analyzing gene sequence datasets. Nowadays, OTU is generally used in many contexts. In most cases, it refers to clusters of uncultivable microorganisms which are grouped according to their DNA sequence similarity. This similarity is detected by a specific taxonomic marker gene. In other words, due to the absence of traditional biological classification systems, OTUs are used as pragmatic substituted terms for microbial individuals at different taxonomic levels.
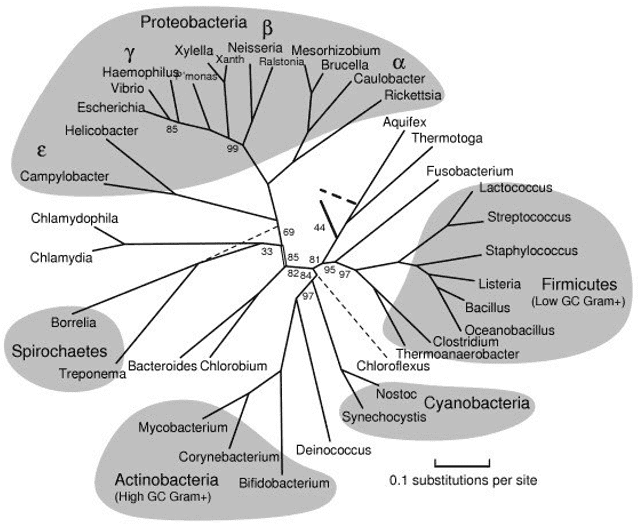
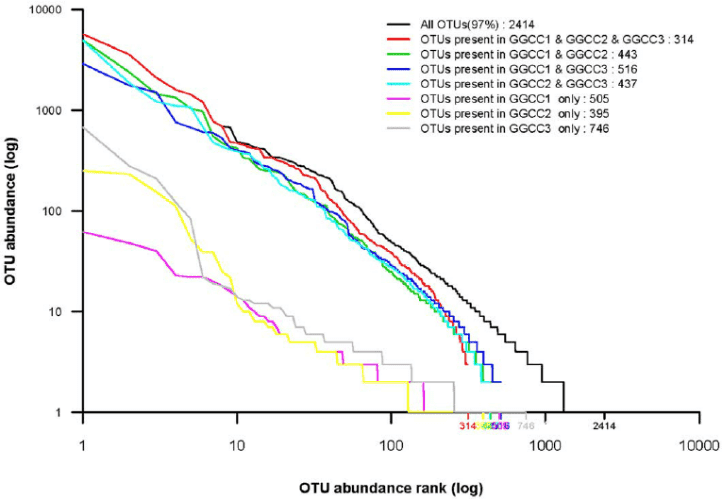
Sequences can be clustered according to similarity. As set by the researcher, the threshold which is usually at 97% similarity determines the clustering of operational taxonomic units. The 16s ribosomal RNA gene is commonly used to study the microbial community. Despite the established algorithms and thresholds for calculating OTUs, some studies demonstrate that microbial OTUs remain generally consistent across habitats and some OTU clustering approaches.
Featured Techniques and Services
Elevate your research with our microbial diversity sequencing, delivering detailed OTU identification and robust bioinformatics for unmatched OTU clustering and diversity analysis.
In constructing taxonomic groups, OTUs that are above the level of the individual need sufficient representation of different polymorphic forms. For instance, when a genus is compared with another genus, they should be represented by species under their respective genera. Likewise, when families are being compared, they should be represented by various genera. OTUs can be visualized by utilizing taxa summaries which depicts relative abundance of the taxa in the set of samples. This provides an efficient way to identify which samples are outliers being notably different from the others. This will also help to visually identify the differences between and among samples.
In microbial research, operational taxonomic units are used to analyse gene sequence data sets obtained through the use of modern sequencing technologies such as metagenomic sequencing and targeted next generation sequencing. Usually, the results generated from the analysis are used in identifying and detecting microorganisms, as well as in developing treatments against disease-causing microbes.
Reference
- He, Y., Caporaso, J., Jiang, X., et al. Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome, 2015, 3 (34).