Application of Rank Abundance Curve in Microbiome Study
The microbiome consists of all microorganisms that exist in a particular microbial community. In a sense, it is the collection of the genetic material of all microbes such as bacteria, archaea, and viruses that live in a specific ecosystem. The study of microbiomes is essential in determining species richness and relative abundances of member microorganisms in a microbial community, correlate it with the mechanisms that control biogeochemical processes in the particular community; and ultimately create a predictive model of the interactions in the microbial community. One of the most commonly used mathematical measures in microbiome studies and other ecological analyses is the rank abundance curve.
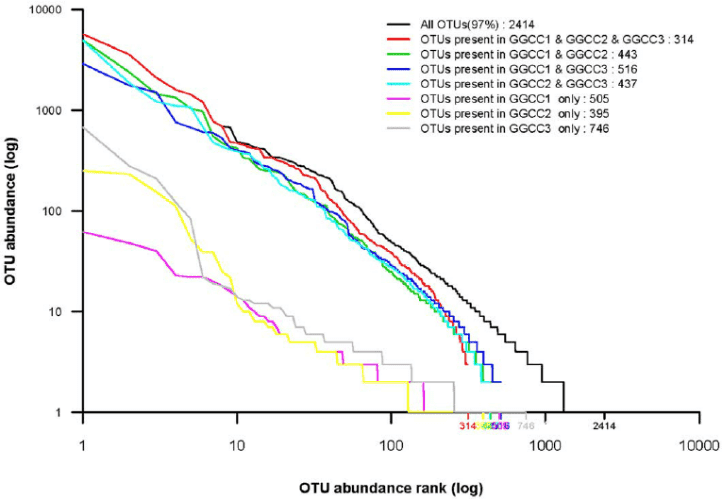 Figure 1. Rank abundance analysis of the different bacterial community groups. (Wu, 2012)
Figure 1. Rank abundance analysis of the different bacterial community groups. (Wu, 2012)
The rank abundance curve or the Whittaker plot is a computational measure of relative species abundance, species richness, and species evenness. It is presented as a graph showing the abundance rank (x-axis) and the relative abundance (y-axis). Species richness is the number of species that were rank while the species evenness is the slope of the line that fits the curve. Species richness often provides an idea of the changes in the number of species from the environmental microbiome. Rank abundance curves provide essential information that supports species richness and community complexity. With the help of this tool, scientists could predict changes in community patterns in response to environmental changes. Rank abundance curves are also used to analyze microbial communities.
Microbial communities, despite them being relatively small, drive major ecological processes in different ecosystems. Studying the behavior of microbial communities help in fully understanding their contribution to the environment at a larger scale. The members of microbial communities or the species richness tend to depend on the changes in the environment, such as availability of resources, temperature, solute concentration, pressure, among others. In terms of species richness and species evenness, microbial communities have varying numbers depending on the changes in the ecosystem and their relative response to these changes. Therefore, the measure of rank abundance in particular microbiomes is essential to predict such changes that could affect the processes occurring in the community.
Some studies are dedicated to profiling the community composition of microbiomes such as the human microbiome. It is known that the human microbiome affects the health of the individual therefore, studying the conditions that could affect the microbiome is important in determining changes that could lead to the development of diseases. Microbial biogeography or the special and temporal patterns of biological diversities in a particular environment is used to determine the presence of microbial groups and their distribution in the community. In the human body, there are particular cells and organ systems that come in contact with different species of microorganisms (niches). Each niche serves as a unique space for the survival and interaction of microbes. Changes in these niches could possibly lead to diseases. Characterizing the human microbial communities by means of high throughput sequencing technologies, such as 16S rRNA gene sequencing, provide information on the diversity and abundance of microbial species in the body.
Aside from applications to human health, rank abundance curves are also used to characterize other microbial communities such as soil microbiomes. The species richness, distribution, and abundance are important measures to determine the conditions in the soil, whether it can be used for agricultural or other applications. General, rank abundance curves help in determining the overall community mechanics and capacity of microbial communities in terms of contributing to biogeochemical processes in nature.
References
- Avolio ML, Carroll IT, Collins SL, et al. A comprehensive approach to analyzing community dynamics using rank abundance curves. Ecospheres. 2019. 10(10).
- Saeedghalati M, Farahpour F, Budeus B, et al. Quantitative comparison of abundance structures of generalized communities: from B-cell receptor repertoires to microbiomes. PLoS computational biology. 2017, 13(1):e1005362.
- Zuo Y, Xie W, Pang Y, et al. Bacterial community composition in the gut content of Lampetra japonica revealed by 16S rRNA gene pyrosequencing. Plos one. 2017, 12(12):e0188919.
- Zhou Y, Gao H, Mihindukulasuriya KA, et al. Biogeography of the ecosystems of the healthy human body. Genome biology. 2013, 14(1):R1.
- Wu S, Wang G, Angert ER, et al. Composition, diversity, and origin of the bacterial community in grass carp intestine. PloS one.2012, 7(2):e30440.