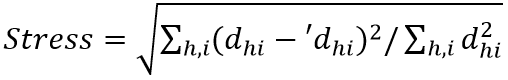The study of the evolutionary relationship between different species of microorganisms is called microbial phylogenetics. The adaptation of the microbial system has redefined the molecular approach to microbial phylogenetic analysis. In describing the reproductive path of species in the past, phylogenetic research examines the evolutionary relationships between organisms thus making it a critical base for microbial research. Developing stable phylogenetic trees is an important step in the characterization of new pathogens and in the development of new biomedicine.
Carl Woese pioneered the molecular approach to phylogenetic research in the 1970s and contributed to the three-domain system (Archaea, Bacteria, Eukarya). The scientific breakthrough in modern molecular biology and the exponential development in computer science have led to a flood of knowledge on nucleic acid sequences, bioinformatics instruments, and methods in phylogenetic inference. In microbiology and the new fields of comparative genomics and phylogenetics, phylogenetic research has played a key role.
Metagenomes and Microbial Phylogeny
Metagenomic data, the cumulative genetic information extracted from an environmental specimen, can be used to research the phylogenetic background of microbial diversity and sample a large variety of genes simultaneously. It is also being used to revolutionize research into gastrointestinal microbes. This highlights the importance of phylogeny to microbiology for classifying behaviors carried out by an ancestral line of animals. The production of reference databases would also support the phylogenetic classification of metagenomic records as more sets of genes are sequenced.
Evolution on Establishing Microbial Phylogeny
The accurate sorting of microorganisms and their relationships with each other is a fundamental process. Prokaryotic organisms are arranged into groups based on morphological or biochemical properties, by early experiments. The primary basis for classifying prokaryotic organisms is the process of generating phylogenetic relations. Phylogenetic studies, specifically the sequence properties of small subunit ribosomal RNA (SSU rRNA), have been improved by molecular analysis. SSU rRNA is an important tool for deciding the evolutionary relationship between species due to its uniform presence, the occurrence of both highly preserved and dynamic regions of the molecule makes. The primary basis for classifying prokaryotic organisms is the process of generating phylogenetic relations.
More recently, new microbiology areas, including comparative genomics and phylogenomics, need extensive knowledge and understanding of phylogenetic analysis and computing skills to work with the large-scale data involved. Phylogenetic research techniques and related computer science resources provide the task with precision, reliability, and availability. The general phylogenetic study of molecular sequences has four stages: (1) selection of a suitable molecule or molecules (phylogenetic marker), (2) acquisition of molecular sequences, (3) multiple sequence alignment (MSA), and (4) phylogenetic treeing and evaluation.
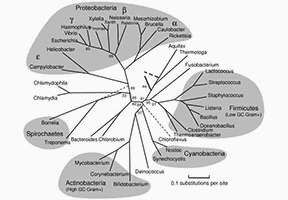
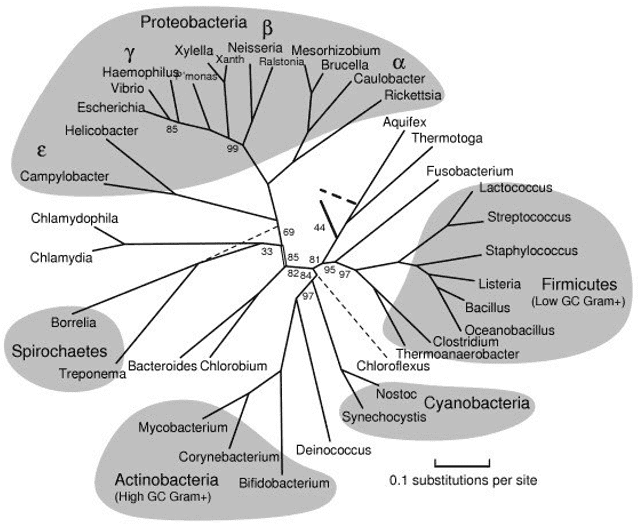 Figure 1. A rooted phylogenetic tree of microorganisms computed from representative proteins (Marsha, 2005)
Figure 1. A rooted phylogenetic tree of microorganisms computed from representative proteins (Marsha, 2005)
Microbial Phylogenetic Tree
There is no official classification of prokaryotes. The most commonly accepted division of the prokaryotes in two "sub-kingdoms" or "domains" (Bacteria and Archaea) and the classification of their species is primarily based on 16S rRNA sequence comparison. Some phyla based on a thorough clade of 16S rRNA are presently embodied only by one or a few species.
References
- Marshall B., Goldberg D.. Automatic selection of representative proteins for bacterial phylogeny. BMC evolutionary biology. 2005, 5. 34.
- Kembel, S.W.. The Phylogenetic Diversity of Metagenomes, PLoS One, 2011, 6, e23214.
- Walker, A. Phylogeny, culturing, and metagenomics of the human gut microbiota, Trends in Microbiology, 2014, 22, pp. 267-274.
- Wu, M. Eisen, J.A.. A simple, fast, and accurate method of phylogenomic inference, Genome Biology, 2008, 9, R151.