Fragment analysis is a form of genetic analysis containing a set of procedures in which fluorescently labeled, capillary electrophoresis isolated and sized DNA fragments are compared to an internal pattern.
Genetic analyzers based on capillary electrophoresis can conduct Sanger sequencing as well as fragment analysis. Fragment analysis can supply sizing, relative quantitation, and genotyping information utilizing fluorescently labeled DNA fragments developed by PCR using primers optimized for a particular DNA target, in relation to Sanger sequencing. Such data helps scientists to classify variations in alleles, homo- and heterozygosity, chimerism, combinations of samples, and inheritance. Fragment research helps a wide range of uses, such as cell line authentication, reliability of CRISPR-Cas9 genome editing assessment, evaluation of microsatellite markers, genotyping of SNP, and more. Fragment analysis has a short processing time and is cost-effective, with strong accuracy and specificity.
Advantages
- Multiplexing - By marking locus-specific primers with various colored dyes, alleles for overlapping loci are differentiated; because of this capacity, fragment analysis allows the assessment of more than 20 loci in a single reaction.
- Sensitivity - Fragments that vary from only one base pair only are precisely sized
- Simple preparation - No DNA cleanup is essential (in contrast to sequencing)
- Easy data analysis - Technology for genetic evaluation simplifies data analysis
- Independent method - does not need the information on the fragment sequence
- Relative quantitation information - From measurements of peak strength
- Automated workflows - Capacity to perform loads of specimens of DNA fragments in a day
- High throughput - can be performed using capillary arrays
Analysis Workflow
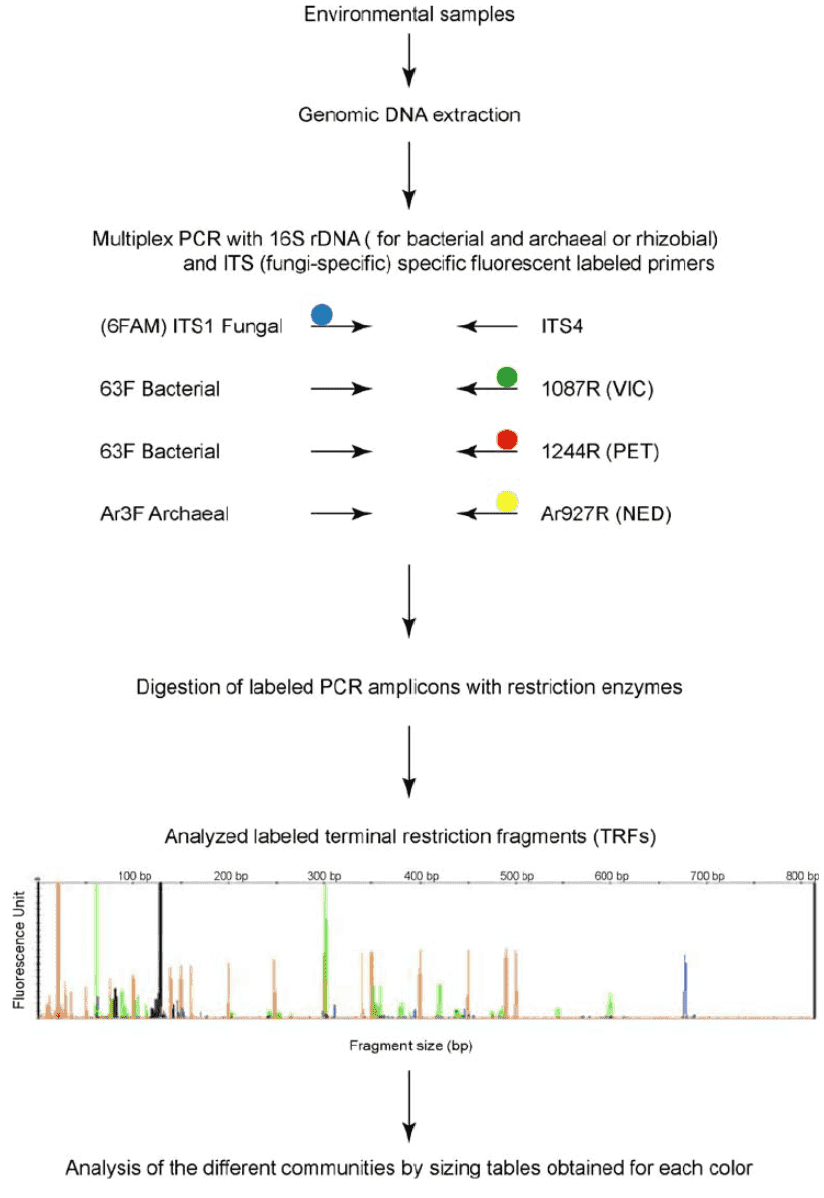 Figure 1. Outline of the procedure for microbial gene fragment analysis by multiplex TRFLP (M-TRFLP) procedure. (Schütte, 2008)
Figure 1. Outline of the procedure for microbial gene fragment analysis by multiplex TRFLP (M-TRFLP) procedure. (Schütte, 2008)
Four main procedures are used in the DNA fragment analysis workflow: DNA isolation, PCR amplification, capillary electrophoresis, and data analysis.
A crucial first step in the experimental process of DNA fragment analysis is DNA extraction. The overall performance, consistency, and size of the PCR product can be greatly influenced by the sample's own attributes and the process selected for the extraction and purification of nucleic acid. Depending on the source or tissue type, how the specimen was collected from its source, and how the specimen was treated or processed prior to extraction, optimal approaches can differ.
Primers that surround the area of interest need to be built to conduct fragment analysis on a CE device. Fluorescent dyes are added to the primers, and before electrophoresis, the fragments are amplified by PCR.
To plan for capillary electrophoresis, in order to reliably identify dye-labeled primers, a spectral calibration with the associating matrix standard for the chosen category of dyes must be done on the genetic analyzer. Until continuing with electrophoresis, any unknown specimen is mixed with the size norm and formamide. Standards of size enable sample peaks to be sized and adjust for differences in injection.
During capillary electrophoresis, the PCR products are electrokinetically inserted into polymer-filled capillaries. High voltage is employed to distinguish the fluorescent DNA fragments by size and a laser/camera device detects them.
Data analysis software produces a separation profile, measures the fragment sizes correctly, and decides the microsatellite alleles in the specimen.
References
- Guo J, Cole JR, Zhang Q, et al. Microbial community analysis with ribosomal gene fragments from shotgun metagenomes. Applied and Environmental Microbiology. 2016 Jan 1;82(1).
- Huang Y, Gilna P, Li W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics. 2009 May 15;25(10).
- Tettelin H, Feldblyum T. Bacterial genome sequencing. Molecular Epidemiology of Microorganisms. 2009.
- Schütte UM, Abdo Z, Bent SJ, et al. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Applied microbiology and biotechnology. 2008 Sep;80(3).







