Not all genes are active at all times. DNA methylation is a common epigenetic modification used by cells to regulate gene expression. In recent decades, researchers have discovered that methylation is involved in many cellular processes, including genomic imprinting, embryonic development, gene suppression, carcinogenesis, X-chromosome inactivation, and preservation of chromosome stability. And abnormal DNA methylation has been proposed to be linked to several adverse outcomes, such as human disease.
DNA is the combinations of four nucleotides, including cytosine, guanine, thymine, and adenine. DNA methylation represents the addition of a methyl (CH3) group to the DNA strand, often to the 5-carbon of the cytosine ring resulting in 5-methylcytosine (5-mC). DNA methylation often modifies the function of the genes and affects gene expression. Methylation is sparse but global in mammal DNA, especially in CpG sites across the entire genome. In somatic cells, 5-mC occurs almost exclusively in CpG sites, while in embryonic stem cells, a substantial amount of 5-mC is observed in non-CpG contexts.
How are genes methylated/demethylated?
The conversion of cytosine bases to 5-mC is catalyzed by DNA methyltransferases (DNMTs). Different DNMTs (DNMT1, DNMT3a, and DNMT3b) work together either as maintenance DNMTs that copy the methylation patterns or as de novo DNMTs that establish new methylation patterns.
DNA demethylation refers to the removal of a methyl group from DNA sequences, which is necessary for epigenetic reprogramming of genes and is directly involved in many disease mechanisms such as tumor progression. Passive DNA demethylation usually occurs on newly synthesized DNA strands during replication rounds. Active DNA demethylation often takes place via the ten-eleven translocation (TET) family of 5-mC hydroxylases. These proteins bind to CpG rich regions to prevent undesired methyltransferase activity, thereby promoting DNA demethylation.
DNA methylation and gene expression
For many years, DNA methylation was believed to play a role in repressing gene expression. In the 80s, it was reported that promoters hypermethylation correlated with reduced expression levels of downstream genes, perhaps caused by blocking the promoters at which activating transcription factors should bind. Paradoxically, DNA methylation is also linked with gene activation, when it takes place within transcribed regions.
Proper DNA methylation is important for the regulation of gene expression. Evidence of this has been found that methylation near gene promoters varies considerably depending on cell type. Additionally, overall methylation levels of particular promotes are similar in individual humans, while there is a significant difference in overall and specific methylation levels between different tissue types and between samples.
Differentially methylated regions (DMRs) are DNA sequences that have distinct methylation status between samples. Genome-wide methylation profiling, such as whole genome bisulfite sequencing, reduced representation bisulfite sequencing (RRBS), EpiTYPER DNA methylation analysis, and MeDIP sequencing, can be performed to identify DMRs and reveal functional regions that are involved in gene transcriptional regulation.
DNA methylation and disease
Given that the vital role of DNA methylation in gene expression, cell differentiation, and embryonic development, it seems that disturbed methylation may give rise to adverse outcomes, such as various human diseases. Indeed, researchers have found that methylation abnormalities may be associated with diseases such as cancer, lupus, muscular dystrophy, birth defects, etc. Epigenetic therapy is a relatively new treatment that focuses on changes in DNA expression. The current epigenetic cancer therapy primarily involves inhibitors of DNA demethylation and histone deacetylation, methylation, and demethylation.
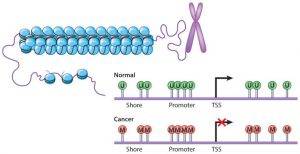
Figure 1. DNA methylation changes in cancer cells (Ahuja N. et al. 2016). Promoter CpG islands in normal cells (top) often lack CpG site DNA methylation ( green U lollipops). In cancer cells (bottom), many genes gain DNA methylation in promoter CpG islands (red M lollipops) with an accompanying repressive chromatin landscape and abnormal gene silencing.
References:
- Ahuja N, Sharma A R, Baylin S B. Epigenetic therapeutics: a new weapon in the war against cancer. Annual review of medicine, 2016, 67: 73-89.
- Phillips T. The role of methylation in gene expression. Nature Education, 2008, 1(1): 116.
- Tirado-Magallanes R, Rebbani K, Lim R, et al. Whole genome DNA methylation: beyond genes silencing. Oncotarget, 2017, 8(3): 5629.


 Sample Submission Guidelines
Sample Submission Guidelines