The vital roles played by small RNA molecules in gene regulation and a myriad of cellular processes are undeniably essential within the realms of biological sciences. This extensive review encompasses a deep exploration into the complex nature of small RNA. We examine its definition, the diversity of its types, its multifaceted functions, as well as its intricate regulation mechanisms. The review also discusses the state-of-the-art advances in small RNA sequencing technologies, reflecting the continuous evolution and growth of this critical field in biotechnology.
Small RNA Definition
Small RNAs constitute a collection of non-coding RNA molecules, typically spanning between 20 to 30 nucleotides in length. Unlike their counterpart messenger RNAs (mRNAs), which function as protein synthesis templates, small RNAs do not have the capacity to encode proteins. Their primary function lies in controlling gene expression at the stage that follows transcription (post-transcriptional level). These molecules are involved in a vast array of biological processes encompassing development, differentiation, immune response, and stress adaptation.
Originating from a diverse set of genomic regions- intergenic regions, introns, exons, and repeated sequences included- small RNAs are transcribed by RNA polymerase II or III, after which they undergo further processing to transform into mature functional molecules. Depending on their subclass, the biogenesis pathways of these small RNAs can range from Dicer-mediated cleavage, the activity of RNA-dependent RNA polymerase (RdRP), to direct ligation.
Small RNA Types
Small RNAs encompass a heterogeneous group of non-coding RNA molecules, renowned for their integral roles in gene regulation amidst other multifaceted cellular processes. These small RNAs comprise various distinct subclasses, each characterized by their unique biogenesis pathways, functional roles, and regulatory modalities.
MicroRNAs (miRNAs)
Among these, miRNAs garnering considerable scientific attention due to their implications in biological processes and disease progression. miRNAs, typically spanning 21-22 nucleotides in length, originate from more complex precursor transcripts, coined as pri-miRNAs. The journey of miRNAs from being a transcript to a mature molecule is multi-staged, commencing with transcription by RNA polymerase II, leading to the generation of pri-miRNAs. The process further progressed by the enzyme-like chaperones Drosha and Dicer, results in the production of mature miRNAs. Subsequently, these miRNAs are loaded into the RNA-induced silencing complex (RISC). This interaction equips miRNAs with the ability to bind to target mRNAs through partial complementarity, causing mRNA degradation or suppressing its translation.
miRNAs play instrumental roles in mediating gene expression and partake in a myriad of biological spheres, including but not limited to, organismal development, cellular differentiation, programmed cell death, apoptosis, and metabolic regulation. Observations reveal that any irregularities in miRNA expression are found to be inextricably linked to a vast array of pathologies, encompassing cancer, cardiovascular diseases, and a plethora of neurological disorders.
Small Interfering RNAs (siRNAs)
The siRNAs represent another primary category of small RNAs that contribute to gene silencing by utilising the RNA interference (RNAi) routes. These siRNAs usually range between 20-25 nucleotides in length and are derived from long double-stranded RNA molecules. The biogenesis of siRNAs involves the Dicer enzyme cleaving dsRNA, thereby generating short siRNA duplexes that are laterally uploaded onto RISC. The guide siRNA strand directs RISC towards complementary mRNA sequences, triggering mRNA cleavage or translation repression.
SiRNAs play a core role in defense against viral attacks, in transposon silencing, and in regulating endogenous gene expression. Their utility has been recognized in gene knockdown during functional genomic studies, and in numerous therapeutic applications.
Piwi-Interacting RNAs (piRNAs)
piRNAs comprise a distinctive category of small RNAs, the expression of which is primarily confined to germline tissues. Generally measuring between 24 and 32 nucleotides, these piRNAs form associations with the specialized Piwi subgroup residing within the larger Argonaute protein family. Their pivotal roles span a range of biological regulations, incorporating transposon silencing, fortification of the genome, and governance of epigenetic phenomena in germ cells.
Contrary to the standard route of small RNA generation, the process of piRNA biogenesis bypasses the requirement for Dicer enzyme, instead being directed by the combined action of the PIWI-piRNA complex. Activated by piRNA guidance, PIWI proteins proceed to silence transposable elements – their action either results in the severing of the transposable elements’ transcripts or instigates the transformation into heterochromatin. This dual mechanism underpinning genetic regulation underscores piRNA’s essential contribution within a wider genetic and epigenetic context.
Small Nucleolar RNAs (snoRNAs)
snoRNAs constitute a significant class of small RNAs that are generally confined to the nucleolus, where they exercise particular influence on the genesis and modification of ribosomal RNA (rRNA) and small nuclear RNA (snRNA). Standard snoRNAs range in length from 60-300 nucleotides and can be categorically divided into two major families based on their conserved sequence motifs: the box C/D snoRNAs and the box H/ACA snoRNAs.
Strikingly, box C/D snoRNAs serve as guides for the 2′-O-methylation of targeted RNA molecules, while the responsibility of box H/ACA snoRNAs is to steer pseudouridylation. These RNA modifications play pivotal roles in fostering the assembly of ribosomes, maintaining RNA stability and safeguarding the accuracy of translation – all crucial to cellular functions.
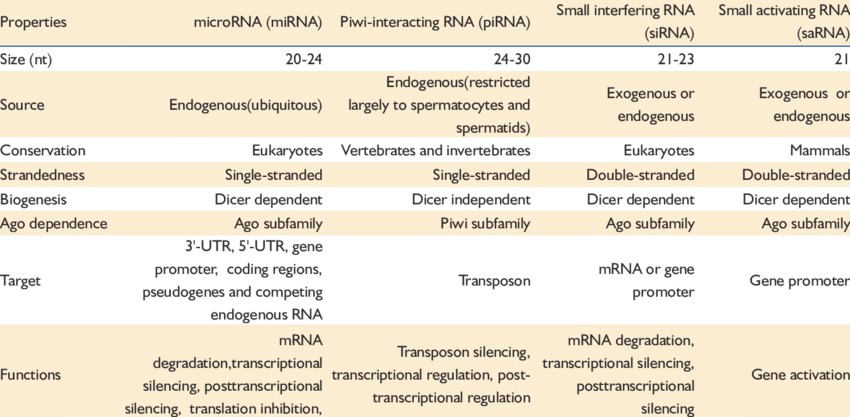
Classes of small RNA. (Ji Wang et al,. 2012)
Small RNA vs. miRNA
| Characteristic | Small RNA | MicroRNA (miRNA) |
|---|---|---|
| Length | Typically 20-30 nucleotides | Typically 21-22 nucleotides |
| Biogenesis | Derived from various biogenesis pathways | Derived from pri-miRNA precursors and processed by Drosha and Dicer |
| Function | Diverse functions including gene regulation, chromatin remodeling, and genome defense | Primarily involved in post-transcriptional gene regulation |
| Regulatory Mechanism | Guides effector complexes to target nucleic acids for silencing or modification | Binds to target mRNAs through complementary base pairing, resulting in translational repression or mRNA degradation |
| Processing Steps | Varied processing steps depending on the specific class of small RNA | Cleavage by Drosha and Dicer to generate mature miRNAs |
| Association with RISC | May or may not associate with RNA-induced silencing complex (RISC) | Incorporated into RISC to guide target mRNA recognition |
| Target Specificity | Targets diverse nucleic acids including mRNAs, other non-coding RNAs, and DNA | Generally targets mRNAs through partial sequence complementarity, leading to mRNA destabilization or translational inhibition |
| Cellular Localization | Found in various cellular compartments including the nucleus and cytoplasm | Predominantly located in the cytoplasm |
| Expression Regulation | Regulated at the transcriptional and post-transcriptional levels | Transcriptionally regulated and subject to post-transcriptional processing |
| Diversity | Includes various classes such as siRNAs, piRNAs, and snoRNAs | Represents a specific class of small RNA with defined biogenesis and function |
Small RNA Function
Pervading a myriad of cellular processes, small RNAs undertake key roles, contributing not only to gene regulation, chromatin architecture, and genome defense, but also orchestrating a panoply of biological pathways. In this discussion, we attempt to uncover the multifaceted duties of small RNAs, illuminating examples curated from established literature. These portray the manifold mechanisms by which small RNAs regulate cellular processes, underlining their biological significance and implications in disease development. Unveiling the intricate responsibilities of small RNas can provide a deeper understanding of complex gene regulation networks and prompt the exploration of therapeutic approaches targeting small RNA-operated courses.
Small RNAs, notably miRNAs, are recognized for their pivotal involvement in post-transcriptional gene regulation. Typically, miRNAs establish binding interactions with the 3′ untranslated region (UTR) of target mRNAs, thereby instigating translational suppression or mRNA degradation. A prominent illustration of this regulatory mechanism is exemplified by the let-7 miRNA family, which exercises control over genes governing cell proliferation and differentiation. Perturbation in the expression levels of let-7 has been linked to a spectrum of malignancies, encompassing lung cancer and breast cancer, underscoring its significance in pathological contexts.
Regarding Chromatin Remodelling, small RNAs are instrumental in shaping chromatin and thereby controlling gene expression. siRNAs are key actors in heterochromatin assembly and transcriptional gene silencing. Demonstrating this, in fission yeast, siRNAs shepherd the RNA-induced transcriptional silencing (RITS) complex towards specific loci, initiating heterochromatin formation and gene suppression.
In Genome Defense, small RNAs form a vital constituent of host protection mechanisms against intrusive nucleic acids like viruses and transposons. For example, plants generate siRNAs in response to viral invasion, targeting the viral RNA for decimation through the RNAi pathway. Likewise, in animal systems, the piRNA pathway mitigates transposable elements to maintain genome stability in germ cells.
Small RNAs also lead pivotal roles throughout various Developmental Processes such as cellular differentiation, embryonic evolution, and organogenesis. miRNAs have emerged as chief regulators of developmental timing in animals with the miR-430 family in zebrafish, as an example, directing the maternal-to-zygotic transition by targeting maternal mRNA for degradation.
Lastly, small RNAs are key moderators in Stress Responses, discerning cellular responses to environmental stressors like heat shock, oxidative stress, and nutrient scarcity. In plants, small RNAs, including miRNAs and siRNAs, regulate stress-responsive genes contributing to biotic and abiotic stress adaptation. For instance, the Arabidopsis miR398 modulates the expression of the copper/zinc superoxide dismutases (CSDs) genes, critical players in oxidative stress resistance.
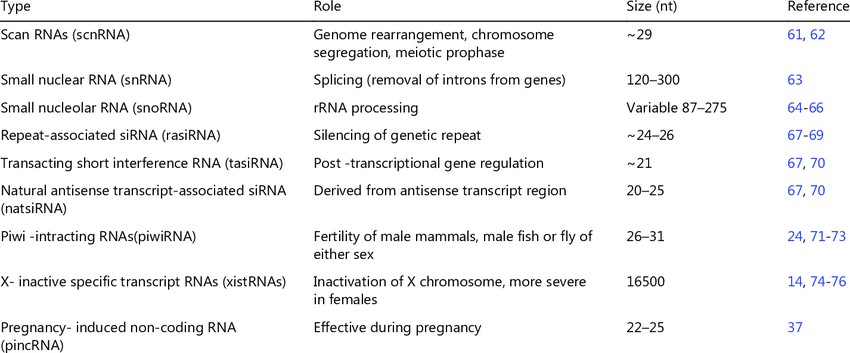
List of small RNAs and their function (Sunny Yadav et al,. 2017)
Small RNA Gene Regulation
Small RNA-mediated gene regulation stands as a cornerstone process, intricately controlling gene expression across multiple tiers, encompassing transcriptional, post-transcriptional, and epigenetic modalities. Within this regulatory framework, diverse classes of small RNAs, including miRNAs, siRNAs, and piRNAs, wield their influence through distinct mechanisms, selectively targeting various facets of the gene expression apparatus. This intricate orchestration underscores the nuanced and multifaceted nature of small RNA-mediated gene regulation, elucidating its pivotal role in governing cellular dynamics and phenotype determination.
Transcriptional Gene Silencing: The intricate orchestration of gene expression finds a pivotal mechanism in small RNAs, notably siRNAs and piRNAs, which wield the capacity to instigate transcriptional gene silencing by navigating chromatin-modifying complexes to precise genomic sites. In plants and fungi, siRNAs spearhead the induction of DNA methylation and histone modifications at targeted loci, thereby precipitating the formation of heterochromatin and subsequent transcriptional repression. Similarly, within the animal kingdom, piRNAs form complexes with PIWI proteins, a partnership designed to silence transposable elements and repetitive sequences within the germline by eliciting heterochromatin formation. These intricate regulatory pathways underscore the multifaceted interplay between small RNAs and chromatin dynamics, delineating their pivotal roles in orchestrating gene expression landscapes across diverse biological contexts.
Post-Transcriptional Regulation: miRNAs emerge as pivotal orchestrators of post-transcriptional gene expression in both animals and plants. Their modus operandi often involves binding to the 3′ untranslated region (UTR) of target mRNAs, eliciting translational repression or mRNA degradation through partial complementarity. Across diverse biological contexts, miRNAs assume crucial roles in fundamental processes such as development, differentiation, and stress responses. However, perturbations in miRNA-mediated gene regulation have been implicated in a spectrum of diseases, ranging from cancer to neurodegenerative disorders. This underscores the profound impact of miRNAs in fine-tuning gene expression dynamics and highlights their significance in both health and disease states.
Epigenetic modifications, a critical aspect of genetic regulation, are facilitated by the guidance of small RNAs directing chromatin-modifying enzymes to precise genomic sites. Within the animal kingdom, pivotal players such as piRNAs undertake the responsibility of orchestrating DNA methylation patterns within the germline, thus safeguarding genomic stability and fidelity. Similarly, in the realm of plants, a diverse array of small RNAs comprising siRNAs and miRNAs exert their influence, modulating DNA methylation dynamics and histone modifications, thereby intricately sculpting gene expression profiles. This intricate interplay underscores the profound impact of small RNAs in shaping the epigenetic landscape across diverse biological systems.
Regulation of Transposable Elements: In the realm of genetic regulation, the management of transposable elements (TEs) and repetitive sequences within the genome stands as a critical endeavor. Small RNAs emerge as pivotal regulators in this process, orchestrating the suppression of TE activity with precision. Notably, siRNAs and piRNAs assume distinct roles in TE silencing within the botanical and zoological domains, respectively. The significance of this regulatory mechanism is underscored by its pivotal role in safeguarding genomic stability and preserving the integrity of adjacent genetic loci. The failure to uphold TE silencing poses a notable risk, potentially precipitating genomic instability and the aberrant expression of proximal genes.
Response to Environmental Stimuli: In the intricate web of cellular responses to environmental cues, small RNAs emerge as crucial orchestrators. Their involvement spans the modulation of gene expression in response to various stimuli, encompassing stressors and alterations in nutrient availability. Through their regulatory prowess, small RNAs finely tune the expression of stress-responsive genes, enabling cells to adeptly navigate shifting environmental landscapes. However, the perturbation of small RNA-mediated gene regulation poses a significant threat, jeopardizing the cellular capacity for stress tolerance and potentially precipitating pathological states. Thus, the nuanced interplay orchestrated by small RNAs underscores their indispensable role in cellular adaptation and resilience amidst environmental challenges.
Small RNA sequencing
Small RNA sequencing, commonly referred to as small RNA-seq, represents a robust methodology employed for the systematic examination and quantification of diminutive RNA species within a given biological specimen. This sophisticated approach empowers investigators to conduct thorough investigations into the intricacies of the small RNA transcriptome, (miRNAs, siRNAs, piRNAs, alongside various other pivotal regulatory RNA entities.
Services you may interested in
Applications of Small RNA-seq
Small RNA sequencing stands as a cornerstone in both fundamental and applied research realms, boasting a myriad of applications that propel scientific inquiry forward. From unearthing novel small RNA entities to delineating their expression profiles across diverse tissues and developmental stages, its utility spans wide. Moreover, small RNA sequencing serves as a potent tool in the quest for disease biomarkers, offering insights crucial for diagnosis and prognosis. Additionally, it unravels the intricate tapestry of small RNA-mediated regulatory networks, shedding light on gene regulation intricacies. Such versatile applications underscore the pivotal role of Small RNA-seq in advancing our comprehension of gene dynamics, disease etiology, and therapeutic modalities. Empowered by the capabilities of high-throughput sequencing technologies, Small RNA-seq continues to spearhead innovations across the biomedical landscape, heralding a new era of discovery and clinical translation.
| Application | Description |
|---|---|
| Discovery of Novel Small RNAs | Small RNA-seq facilitates the discovery of novel small RNA species, including miRNAs, siRNAs, and piRNAs, by capturing and sequencing all small RNA molecules present in a sample. |
| Characterization of Small RNA Profiles | Small RNA-seq enables the comprehensive characterization of small RNA expression profiles in different tissues, developmental stages, and disease conditions. By quantifying the abundance of small RNAs, researchers can identify tissue-specific or disease-associated small RNAs. |
| Identification of Disease Biomarkers | Small RNA-seq has been instrumental in identifying small RNA biomarkers associated with various diseases, including cancer, cardiovascular disorders, and neurological conditions. Differential expression analysis of small RNAs between diseased and healthy samples can reveal potential biomarkers for disease diagnosis, prognosis, and treatment response. |
| Elucidation of Regulatory Networks | Small RNA-seq provides insights into the intricate regulatory networks governed by small RNAs. By identifying target genes regulated by miRNAs and other small RNAs, researchers can elucidate the molecular mechanisms underlying gene expression regulation and cellular processes. |
| Functional Annotation of Small RNAs | Small RNA-seq data can be used for functional annotation of small RNAs, including predicting their target genes and pathways. Integrating small RNA expression profiles with transcriptomic and proteomic data enables the functional annotation of small RNAs and their regulatory roles in biological processes. |
| Investigation of RNA Modifications | Small RNA-seq can be used to study RNA modifications, such as 3′ end modifications and nucleotide editing, which play crucial roles in small RNA stability, biogenesis, and function. By mapping small RNA sequences to the reference genome, researchers can identify and characterize RNA modifications in small RNAs. |
| Drug Discovery and Therapeutic Targeting | Small RNA-seq data can inform drug discovery efforts by identifying small RNAs implicated in disease pathogenesis and potential therapeutic targets. Targeting dysregulated small RNAs with small molecule inhibitors or RNA-based therapeutics holds promise for developing novel treatments for various diseases. |
Challenges and Considerations of Small RNA-seq
While undeniably invaluable, Small RNA sequencing encounters several limitations and hurdles that warrant attention. Chief among these are the challenges associated with detecting low-abundance small RNAs, the potential biases introduced during library preparation and sequencing, and the intricacies inherent in data analysis. Mitigating these challenges necessitates a concerted effort towards optimizing library preparation protocols, refining experimental design strategies, and enhancing the robustness of bioinformatic pipelines. Such endeavors are imperative to ensure the attainment of precise and reproducible results, thereby bolstering the reliability and efficacy of small RNA-seq as a cornerstone technique in biological research.
References:
- Zhang C. Novel functions for small RNA molecules. Curr Opin Mol Ther. 2009 Dec;11(6):641-51. PMID: 20072941; PMCID: PMC3593927.
- Benesova S, Kubista M, Valihrach L. Small RNA-Sequencing: Approaches and Considerations for miRNA Analysis. Diagnostics (Basel). 2021 May 27;11(6):964.
- V. Narry Kim Small RNAs: Classification, Biogenesis, and Function Molecules and Cells 2005
- Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21-44.
- Xiong, Q., Zhang, Y. Small RNA modifications: regulatory molecules and potential applications. J Hematol Oncol 16, 64 (2023).


 Sample Submission Guidelines
Sample Submission Guidelines