What Is Sanger Sequencing?
Sanger sequencing, also known as the “chain termination method” was developed by the British biochemist Frederick Sanger and his colleagues in 1977. This method is designed for determining the sequence of nucleotide bases in a piece of DNA (commonly less than 1,000 bp in length). Sanger sequencing with 99.99% base accuracy is considered the “gold standard” for validating DNA sequences, including those already sequenced through next-generation sequencing (NGS). Sanger sequencing was used in the Human Genome Project to determine the sequences of relatively small fragments of human DNA (900 bp or less). These fragments were used to assemble larger DNA fragments and, eventually, entire chromosomes.
Sanger Sequencing VS NGS
The development of NGS technologies has accelerated genomics research with high speed, high throughput, and powerful solutions. NGS can simultaneously interrogate more than 100 genes at a time and sequence whole genomes with low-input DNA. However, Sanger sequencing remains widely used in the sequencing field as it offers several prominent advantages: (i) cost-efficiency for sequencing single genes and (ii) 99.99% accuracy, especially suitable for verification sequencing for site-directed mutagenesis or cloned inserts.
How Does Sanger Sequencing Work?
In sanger sequencing, a DNA primer complementary to the template DNA (the DNA to be sequenced) is used to be a starting point for DNA synthesis. In the presence of the four deoxynucleotide triphosphates (dNTPs: A, G, C, and T), the polymerase extends the primer by adding the complementary dNTP to the template DNA strand. To determine which nucleotide is incorporated into the chain of nucleotides, four dideoxynucleotide triphosphates (ddNTPs: ddATP, ddGTP, ddCTP, and ddTTP) labeled with a distinct fluorescent dye are used to terminate the synthesis reaction. Compared to dNTPs, ddNTPs are removed an oxygen atom from the ribonucleotide, hence cannot form a link with the next nucleotide. Following synthesis, the reaction products are loaded into four lanes of a single gel depending on the diverse chain-terminating nucleotide and subjected to gel electrophoresis. According to their size, the sequence of the DNA is thus determined.
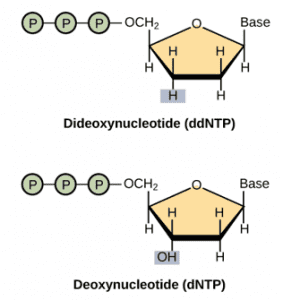
Figure 1. The structure of ddNTP and dNTP.
Image credit: “Whole-genome sequencing: Figure 1,” by OpenStax College, Biology).
Sanger Sequencing Steps
The Sanger sequencing method consists of 6 steps:
(1) The double-stranded DNA (dsDNA) is denatured into two single-stranded DNA (ssDNA).
(2) A primer that corresponds to one end of the sequence is attached.
(3) Four polymerase solutions with four types of dNTPs but only one type of ddNTP are added.
(4) The DNA synthesis reaction initiates and the chain extends until a termination nucleotide is randomly incorporated.
(5) The resulting DNA fragments are denatured into ssDNA.
(6) The denatured fragments are separated by gel electrophoresis and the sequence is determined.
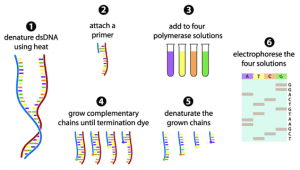
Figure 2. The Sanger sequencing method in 6 steps (adapted from Gauthier 2008).
References:
- Gauthier, Michel G. Simulation of polymer translocation through small channels: A molecular dynamics study and a new Monte Carlo approach. Diss. University of Ottawa (Canada), 2008.
- Sikkema‐Raddatz, Birgit, et al. Targeted next‐generation sequencing can replace Sanger sequencing in clinical diagnostics. Human mutation 34.7 (2013): 1035-1042.


 Sample Submission Guidelines
Sample Submission Guidelines