In the field of genomics, two notable methodologies for DNA sequencing have risen to prominence: Primer Walking Sequencing and Shotgun Sequencing. These techniques, although distinct in their approaches, stand as indispensable instruments for deciphering the intricacies inherent within the genetic code. This discourse endeavors to dissect the nuances of both methodologies, elucidating their fundamental principles, diverse applications, as well as their respective strengths and limitations.
What is Primer Walking Sequencing
Primer walking sequencing stands as a directed approach for deciphering the nucleotide sequence within a designated genomic segment. Unlike the stochastic fragmentation and parallel sequencing employed in Shotgun Sequencing, primer walking initiates from a predetermined DNA sequence contiguous to the target region. This method entails the incremental elongation of DNA fragments originating from the established sequence, facilitated by a succession of overlapping primers. Each primer is meticulously crafted to anneal to the terminus of the preceding sequenced fragment, thereby orchestrating a sequential advancement along the template DNA strand, akin to a deliberate "walk" through the genetic landscape.
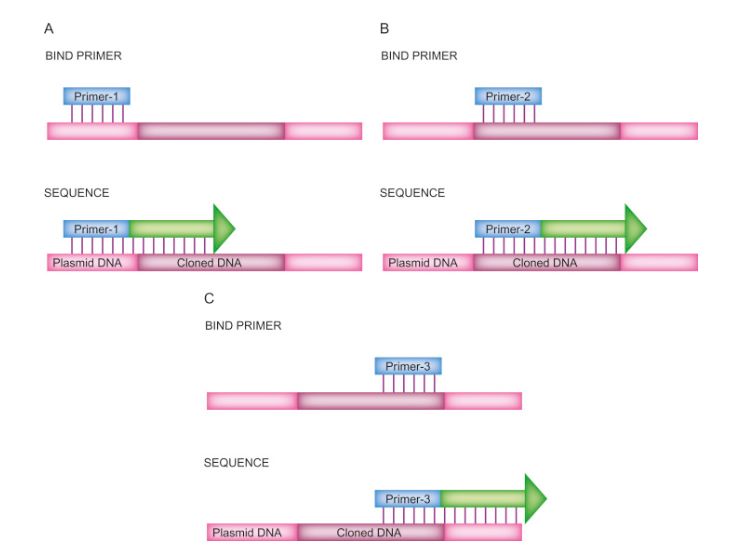
Primer walking is a quicker and easier method of sequencing long stretches of DNA (David P. Clark, Nanette J. Pazdernik, Molecular Biology, 2013)
Application of Primer Walking Sequencing
Targeted Gene Sequencing
Primer walking sequencing is commonly employed for the targeted sequencing of individual genes or specific genomic regions of interest. It is particularly valuable in clinical diagnostics, where the accurate determination of genetic variants within specific genes is essential for disease diagnosis, prognosis, and treatment selection. By selectively amplifying and sequencing the target region, primer walking enables the precise identification of single nucleotide polymorphisms (SNPs), insertions, deletions, and other genetic variations associated with inherited diseases or cancer.
Characterization of Complex Genomic Loci
In research settings, primer walking sequencing is employed to characterize complex genomic loci, including regions with repetitive sequences, structural variations, or epigenetic modifications. These regions often pose challenges for standard sequencing approaches due to their repetitive nature or genomic instability. Primer walking allows researchers to systematically sequence and assemble the entire sequence of such regions, providing valuable insights into their structure, function, and evolutionary conservation.
Validation of Next-Generation Sequencing (NGS) Data
Primer walking sequencing is also used to validate and complement data generated by NGS technologies. While NGS offers high throughput and scalability, it may encounter difficulties in accurately resolving certain genomic regions, such as GC-rich regions or repetitive sequences. Primer walking can be employed to fill gaps, confirm variants, or resolve ambiguities in the NGS data, ensuring the accuracy and completeness of the genomic sequence.
Advantages of Primer Walking Sequencing
High Accuracy and Precision
One of the primary advantages of primer walking sequencing is its high accuracy and precision in determining the nucleotide sequence of the target region. By iteratively extending DNA fragments from a known starting point, primer walking ensures that each nucleotide is accurately determined and confirmed through overlapping sequencing reads. This precision makes it ideal for applications requiring the detection of subtle genetic variations, such as disease-associated mutations or regulatory elements.
Targeted Sequencing
Primer walking sequencing offers the flexibility of targeted sequencing, allowing researchers to focus on specific genomic regions of interest. Unlike shotgun sequencing, which sequences the entire genome indiscriminately, primer walking selectively amplifies and sequences the target region, reducing the sequencing cost and data complexity. This targeted approach is particularly advantageous for clinical diagnostics, where the accurate characterization of disease-related genes is paramount.
De Novo Sequencing of Complex Regions
In addition to targeted sequencing, primer walking can be used for de novo sequencing of complex genomic regions, including regions with repetitive sequences, structural variations, or unknown flanking sequences. By systematically extending DNA fragments from a known starting point, primer walking enables the assembly of contiguous sequences spanning the entire region of interest. This capability is valuable for genome annotation, comparative genomics, and evolutionary studies, where the accurate reconstruction of complex genomic loci is essential.
What is Shotgun Sequencing
Shotgun sequencing represents a high-throughput technique in DNA sequencing, employed for the comprehensive decoding of an organism’s entire genome or a designated genomic segment. In contrast to the targeted methodology of primer walking sequencing, shotgun sequencing adopts a stochastic approach by fragmenting DNA into smaller units, sequencing them individually, and subsequently assembling the fragments into a cohesive sequence through computational algorithms.
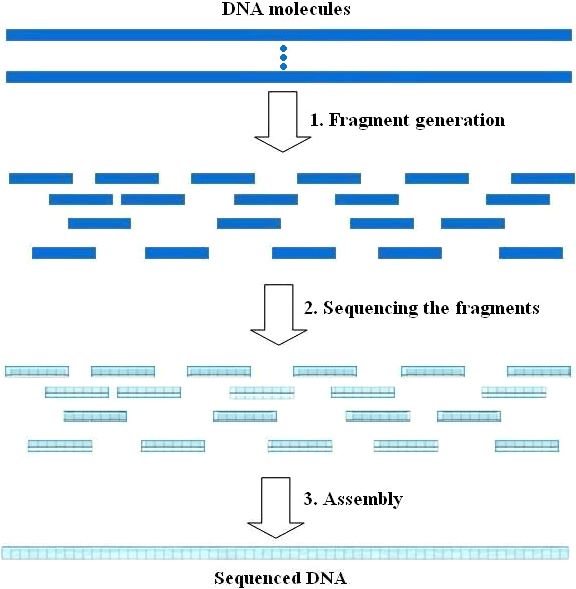
A simple illustration of the shotgun sequencing steps. (REZA POURMOHAMMADI et al,. 2019)
Application of Shotgun Sequencing
Whole Genome Sequencing
One of the foremost applications of shotgun sequencing lies in whole genome sequencing (WGS), wherein the complete genetic blueprint of an organism is deciphered devoid of prior sequence information. This method facilitates a thorough genomic exploration encompassing gene identification, regulatory elements, repetitive sequences, and structural variations spanning the entirety of the genome. WGS finds extensive utility across diverse domains such as research, clinical diagnostics, evolutionary biology, and personalized medicine, serving as a cornerstone for investigations into genetic diversity, disease origins, and population genetics.
Metagenomic Analysis
In addition to its applications in whole genome sequencing, shotgun sequencing finds utility in metagenomic (Metagenomic Shotgun Sequencing) investigations aimed at unraveling the genetic makeup of intricate microbial communities within environmental samples, such as soil, water, or the human gut microbiome. Through the sequencing of DNA fragments extracted from diverse microbial populations, shotgun metagenomics enables the identification and delineation of microbial diversity, functional capacities, and metabolic pathways inherent within a specified ecosystem. This methodology holds broad-ranging implications in fields such as environmental microbiology, biotechnology, and human health, facilitating inquiries into microbial ecology, infectious diseases, and bioremediation strategies.
Comparative Genomics
Within the realm of comparative genomics, shotgun sequencing serves as a pivotal tool for scrutinizing the genomes of diverse organisms or closely allied species, aiming to unravel intricate evolutionary relationships, genomic rearrangements, and adaptive traits. Through the sequencing of multiple genomes and subsequent comparative analyses, researchers endeavor to delineate conserved regions, lineage-specific genes, and genomic disparities correlated with phenotypic variations, speciation phenomena, or environmental adaptations. The field of comparative genomics carries profound implications for evolutionary biology, biodiversity preservation efforts, and the elucidation of intricate biological mechanisms.
Services you may interested in
Advantages and Disadvantages of Shotgun Sequencing
Advantages of Shotgun Sequencing
High Throughput: Shotgun sequencing boasts remarkable high throughput capabilities, facilitating the swift sequencing of expansive genomes or numerous samples concurrently. This scalability renders it well-suited for ambitious genomic endeavors and extensive population studies.
Comprehensive Coverage: Through the random fragmentation of the genome, shotgun sequencing achieves exhaustive coverage of the entire genetic landscape, encompassing repetitive sequences and structural variations. This impartial methodology ensures the identification of elusive variants and genomic rearrangements.
Versatility: Shotgun sequencing exhibits remarkable adaptability, applicable to diverse DNA origins including whole genomes, metagenomic specimens, transcriptomes, and targeted segments. Such versatility renders it invaluable across a broad spectrum of research pursuits spanning various biological domains.
Disadvantages of Shotgun Sequencing
Computational Complexity: The computational demands inherent in assembling shotgun sequencing data into a cohesive genome sequence pose significant challenges, particularly evident in intricate genomes or regions rife with repetitive elements. Assembly errors, misalignments, and genomic lacunae may arise, necessitating meticulous manual curation and validation.
Cost: Despite strides in sequencing technology, the expense associated with shotgun sequencing remains considerable, especially concerning large-scale genomes or projects demanding profound coverage depth. The procurement of sequencing reagents, equipment, and the computational infrastructure for data analysis can act as formidable barriers, constraining accessibility, particularly for research cohorts with constrained financial resources.
Bioinformatics Expertise: Proficiency in bioinformatics and computational biology proves indispensable for the accurate processing, alignment, and interpretation of shotgun sequencing data. Rigorous data management, quality assurance protocols, and adept utilization of bioinformatics resources are paramount for robust genomic scrutiny and coherent interpretation.
Primer Walking vs Shotgun Sequencing
Accuracy and Precision
Primer walking sequencing is renowned for its high accuracy and precision, particularly in regions with complex structures or repetitive sequences. By iteratively extending the sequence from a known starting point, primer walking ensures that each nucleotide is accurately determined.
This precision makes it ideal for applications requiring the detection of SNPs or small insertions/deletions (indels) with utmost confidence.
In contrast, shotgun sequencing, while offering high throughput, may encounter challenges in accurately resolving repetitive regions or distinguishing between homologous sequences. The random fragmentation of DNA leads to overlapping reads, which must be assembled computationally. However, in regions with repetitive elements or sequence duplications, assembly errors or misalignments may occur, compromising the accuracy of the reconstructed sequence.
Throughput and Scalability
Primer walking sequencing, due to its iterative nature and reliance on prior knowledge of adjacent sequences, typically offers lower throughput and scalability compared to shotgun sequencing. It is well-suited for targeted sequencing of individual genes or specific genomic regions but may not be efficient for sequencing entire genomes or multiple samples simultaneously.
On the other hand, shotgun sequencing excels in throughput and scalability, allowing for the rapid sequencing of entire genomes or large datasets of samples. By randomly fragmenting the DNA and sequencing the fragments in parallel, shotgun sequencing enables high-throughput analysis, making it indispensable for large-scale genomic studies and population-level analyses.
Dependency on Prior Knowledge
A notable distinction between primer walking and shotgun sequencing lies in their dependency on prior knowledge of the target sequence. Primer walking requires a contiguous sequence adjacent to the region of interest for primer design and initiation of sequencing. This prerequisite limits its applicability to regions with known flanking sequences, making it less suitable for de novo sequencing or exploration of unknown genomic regions.
In contrast, shotgun sequencing does not rely on prior knowledge of the target sequence, offering greater versatility and applicability to de novo sequencing projects. By randomly fragmenting the DNA, shotgun sequencing can sequence entire genomes or complex regions without the need for prior sequence information, making it indispensable for genome assembly and discovery-driven research.
Computational Complexity
While primer walking sequencing involves relatively simple data analysis, with straightforward interpretation of sequencing results, shotgun sequencing presents challenges in computational complexity. The large volume of short sequencing reads generated by shotgun sequencing requires sophisticated computational algorithms for accurate assembly and analysis. Assembly algorithms must address issues such as read alignment, error correction, and resolution of repetitive sequences to reconstruct the original genome sequence accurately.
Moreover, shotgun sequencing data often necessitates post-assembly analysis for variant detection, genome annotation, and comparative genomics studies, further adding to the computational complexity. Advanced bioinformatics tools and computational resources are essential for processing and interpreting shotgun sequencing data effectively.
Conclusion
In summary, both primer walking sequencing and shotgun sequencing offer distinct advantages and limitations in genomic research. While primer walking excels in accuracy, precision, and targeted sequencing applications, shotgun sequencing provides high throughput, scalability, and versatility for large-scale genome analysis and de novo sequencing projects. The choice between these methods depends on the specific research objectives, the complexity of the genomic region under investigation, and the available resources and expertise.


 Sample Submission Guidelines
Sample Submission Guidelines