Ten years ago, clinicians at several hospitals across the United States began sequencing the genomes of seemingly healthy infants in an attempt to understand how this technology would be able to detect recessive genetic disorders that would not be detected by conventional newborn blood tests.
Recently, a team of researchers from BabySeq, the world’s first clinical randomized trial of newborn genome sequencing, published in the American Journal of Human Genetics (AJHG) the latest results of the project, which found that about 11 percent of the first infants screened for genome sequencing were found to have unanticipated mutations in disease-related genes. Meanwhile, the research team followed the infants and families for three to five years.
The BabySeq Project
The BabySeq project is the first randomized clinical trial of its kind, first initiated in 2013 by a collaboration between BWH and BCH and expanded to Massachusetts General Hospital (MGH) with funding from the National Institutes of Health. The trial is designed to evaluate the utility of genomic sequencing in routine neonatal care. The BabySeq project helps complement and expand existing newborn screening while screening for thousands of diseases that newborns may be at risk for developing in childhood. At the same time, early diagnosis of these diseases can in turn facilitate the development of specific screening, surveillance and treatment protocols, leading to more personalized and preventive healthcare.
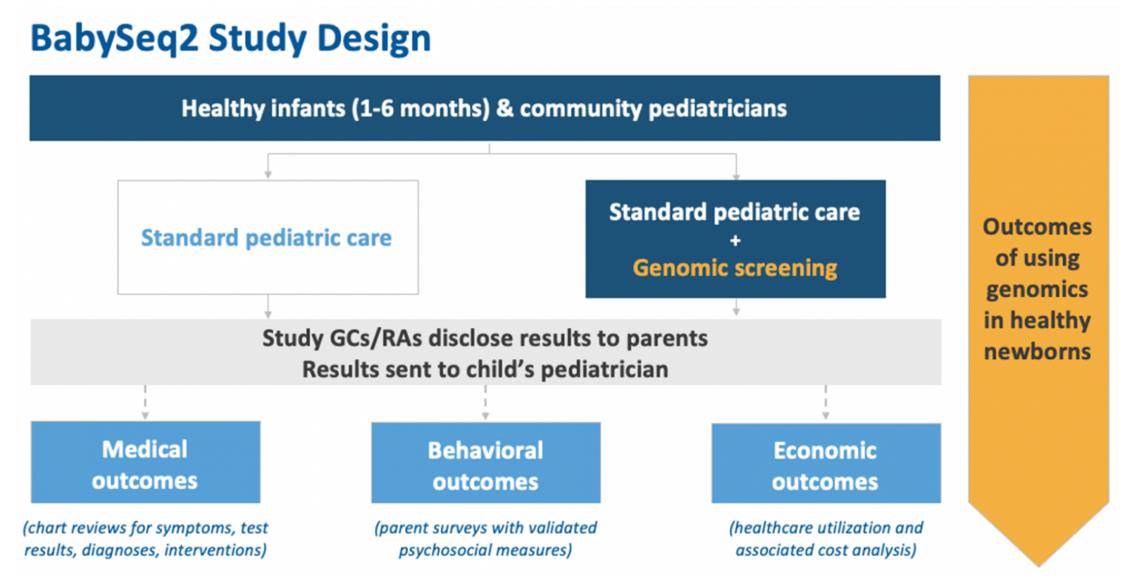
The BabySeq Project form G2P
BabySeq Helps Complement and Expand Existing Newborn Screenings
In previous studies, researchers at Mass General Brigham and Boston Children’s Hospital (BCH) screened the first 159 infants, including 127 healthy infants and 32 infants in intensive care, by comprehensive exome sequencing. Single-gene disease risks (uMDRs) were identified in 10.7% (17) of the infants; 88% of the newborns had at least one recessive carrier variant, which may be relevant to their parents’ future reproductive plans; and 5% had atypical pharmacogenomic variants related to how they dealt with childhood drug use.
In the current report, the team assessed the actionability of each uMDR using the improved Clinical Genome Resource Center (ClinGen) Actionable Semi-Quantitative Measure (CASQM) and found that all of these mutations are medically actionable, meaning that children may benefit from early treatment or monitoring. The research team also created radar plots indicating the extrapolation of the condition, the severity of the condition, the effectiveness of the intervention and the tolerability of the intervention. In addition, the researchers followed the infants for 3-5 years and tracked the medical interventions triggered by these findings.
Results showed that in three infants, uMDR revealed genetic etiologies not anticipated by existing phenotypes, and in the remaining 14 infants, uMDR provided risk stratification for future medical surveillance. In addition, in 13 infants, uMDR prompted screening of at-risk family members. Of interest, three infants whose high-risk mothers were identified as being at high risk for previously identified adult-onset cancers chose to undergo risk-reducing surgery.
Conclusion
Neonatal gene lease sequencing can reveal not only the risk of future disease, but in some cases hidden health conditions that already exist. For example, in a healthy newborn recruited for the study, researchers detected a deleterious mutation in the ELN gene that causes supra-aortic stenosis, which can lead to heart failure if left untreated. During follow-up, a previously unanticipated aortic stenosis was identified in that infant.
The BabySeq project has published several findings on the impact of sequencing in newborns. The project is currently preparing for a Phase II study and is recruiting newborns and their families in multiple cities to include a diverse, nationally representative cohort of families.
As the cost of genetic sequencing has fallen dramatically, the prospect of screening millions of newborns with whole-genome sequencing raises deep concerns about how useful this genetic information really is. Researchers remain divided on whether the benefits of early detection of disease outweigh the added costs and burdens on the health care system, and on the potential psychological impact on families of knowing they carry genes at risk for disease. What is undeniable, however, is that whole-genome sequencing could lead to new breakthroughs in newborn screening, helping clinicians to more comprehensively, efficiently, and accurately identify potential disease risks in newborns, leading to earlier treatment and medical interventions, improved quality of life for newborns and their families, and a lower burden on society.
Reference
- Robert C. Green, Actionability of Unanticipated Monogenic Disease Risks in Newborn Genomic Screening: Findings from the BabySeq Project, The American Journal of Human Genetics (2023). DOI: 10.1016/j.ajhg.2023.05.007.
- https://www.genomes2people.org/research/babyseq/


 Sample Submission Guidelines
Sample Submission Guidelines