The NGS Revolution: Enabling Drug Development and Personalized Medicine
What is Next Generation Sequencing
Next generation sequencing (NGS), also known as massively parallel or deep sequencing, describes a DNA sequencing technology that can sequence an entire human genome within a single day. It has emerged as a revolutionary technology in genomics research. The NGS methods are based on four techniques, including (i) microchip based electrophoretic sequencing, (ii) real-time sequencing, (iii) sequencing by hybridization, and (iv) cyclic-array sequencing. The workflow of NGS experiments is composed of several basic steps, including genomic template preparation, sequencing library preparation, generation of short sequence reads, reads alignment, sequence assembly, and advanced data analysis. Popular NGS platforms include Roche/454 life sciences, Illumina/Solexa, and Ion Torrent.
Services you may interested in
How can NGS Contribute to Drug Development
Multiple mutated genes are found to be associated with disease development and progression. In myeloma, BRAF, NRAS, and KIT are mutation causing genes. Gene therapies that target these genes can reduce metastatic growth. For another example, the breast cancer progression can be estimated based on the difference in read length of CAG repeats due to the intra-tumor genetic heterogeneity of androgen receptor gene. The NGS technology has been considered as a promising approach for pharmaceutical industries and clinical practice, especially in the field of cancer research, since cancers are primarily caused by gene mutations. NGS allows the identification of gene mutations, disease diagnosis and determination of disease progression via biomarker prediction, and genetically stratified clinical trials.
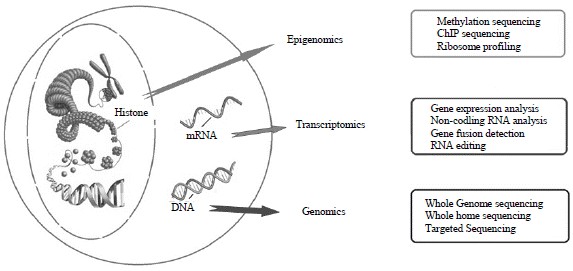 Figure 1. Different approaches of next generation sequencing technologies (Kumar et al. 2017).
Figure 1. Different approaches of next generation sequencing technologies (Kumar et al. 2017).
NGS methods, such as whole genome sequencing, whole exome sequencing, transcriptomic sequencing, and targeted sequencing, can be utilized to perform molecular profiling, so as to discover novel drug targets or biomarkers. While whole genome sequencing detects whole-genome mutations, whole exome sequencing focuses on 1-2% of entire genome. But whole exome sequencing covers more than 95% of exons, which is much higher than whole genome sequencing. Transcriptome sequencing has been carried out to profile mRNA expression analysis and detect non-coding RNAs, such as miRNA, siRNA, piRNA, and lncRNA, which may be potential biomarkers. Targeted panel sequencing is characterized by high depth and high exon coverage, and is quite useful in detecting scarce variants and valuable variants.
Additionally, epigenomic sequencing, such as ChIP-sequencing, ribosome profiling, and bisulfite sequencing, has merged as a powerful tool for drug development and clinical practice. DNA methylation, histone modification, and chromatin remodeling are critical epigenetic mechanisms for gene and non-coding RNA expression. In addition to epigenetic diseases such as Alzheimer's disease, many cancer therapies are based on epigenetic drugs. Epigenetic drugs have several advantages. First, many diseases depend not only on mutations, but chiefly on altered levels of expression of epigenetic modulators. Second, epigenetic component variations have a relatively low toxicity and are well tolerated in normal tissues as long as critical thresholds are not crossed. Third, combination with drugs acting on other mechanisms is very promising since epigenetic drugs leverage different and frequently orthogonal cell processes.
How can NGS Contribute to Personalized Medicine
Next generation sequencing has revolutionized an era of genome sequencing and medical science. It provides faster tools for screening of biomarkers, molecular profiling, and reliable identification of genetic interaction. For complex diseases, such as cancer, NGS is quite useful in identifying the causing factors of specific therapeutic conditions, monitoring disease progression, and determining personalized treatment. Researchers have discovered novel and rare mutations associated with particular therapeutic conditions.
- Tumor markers
Anyone can develop cancer. If they are diagnosed accurately and early using proper approaches, a substantial proportion of cancers can be prevented. A cancer diagnosis can be difficult. First, most cancers are characterized by silence. Second, the patient may be asymptomatic or have symptoms of other noncancerous diseases. Currently, genome-based blood tests are a novel approach to detect tumor markers.
Tumor markers can be discovered by detecting SNPs, which are the most common mutations in the population. The next step is the development of strategies for the identification and utilization of personalized tumor markers by using individual genetic profiles. Thus far, personalized tumor markers have enabled researchers for many aspects, including early cancer detection, screening, diagnosis, prognosis, targeted therapy, therapeutic response, monitoring and recurrence.
- HLA system in the clinical treatment
Pharmacogenomics represents the combination of pharmacology and genomics. It studies how drugs respond differently from person to person based on genomic content, and how genotype-phenotype information can be used in personalized medicine. The major benefits of pharmacogenomics are as follows:
1. The development of drugs to maximize their therapeutic effects and minimize their adverse effects.
2. The ability to design appropriate dosage methods based on patients' genetic profile.
3. Identification of the responders and non-responders for drugs and determination of the risk factors for that drug.
Services you may interested in
Conclusions
The NGS has been applied in biopharmaceutics, polypharmacology, vaccinology, pharmacoepidemiology, etc. For antibody development, NGS data provide a more precise analysis of multiplicity in an antibody library. And NGS can be used to study protein-protein interactions and antibody-antigen binding. But there are also some considerations when performing NGS experiments. For example, the genetic content of tissue can be affected by heterogeneity, such as in tumor samples. The composition varies across samples and may confound disease diagnosis and determination of treatment. Signal may originate from diverse cell types, and in silico optimization and gene list-based approaches have been applied to stratify signals into their respective cell types. Single-cell sequencing has been successfully used to predict treatment response.
You can also refer to our articles "Next-generation sequencing for cancer biomarker discovery" and "Sequencing Technology for Antibody Drug Discovery".
References:
- Kumar G, Chaudhary K K, Misra K, et al. Next-Generation Sequencing for Drug Designing and Development: An Omics Approach for Cancer Treatment. Int. J. Pharmacol, 2017, 13(7): 709-723.
- Lightbody G, Haberland V, Browne F, et al. Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application. Briefings in bioinformatics, 2018, 1: 17.
- Nawab D H. The Pharmaceutical Applications of Next Generation Sequencing in Oncology Drug Designing and Development. Journal of Next Generation Sequencing & Applications, 2015, 2:1.


 Sample Submission Guidelines
Sample Submission Guidelines
