Next-Generation Sequencing for Cancer Biomarker Discovery
What are biomarkers?
Biomarkers are key molecular, chemical or cellular features that can be objectively measured and used to describe biological processes, pathogenic states and responses to treatment. Biomarkers can be disease-related or treatment-related. Disease-related biomarkers are diagnostic (used to determine disease state), prognostic (provide information about potential clinical outcomes, independent of treatment) or predictive (provide information about potential clinical outcomes in response to a specific treatment)
For a biomarker to be acceptable for clinical application, it must have the following characteristics:
1. be easily and consistently detectable in biological fluids, tissues or other biological specimens
2. be rapidly detectable and stable.
3. High sensitivity and specificity.
4. Strong correlation with the phenotype or result of interest.
5. Can be detected by simple, non-invasive and cost-effective tests.
6. No gender-specific
Instead, cancer biomarkers should be specific to the cancer subtype, provide information about the metastatic potential of the cancer and should also be detectable in archived samples such as FFPE (formalin-fixed paraffin-embedded). As cancer is caused by genetic aberrations and is a heterogeneous disease, molecular biomarkers such as genetic variants, gene expression profiles and, in some cases, genomic methylation status, may provide more actionable insights than traditional markers. Sequencing technologies capable of performing millions of reactions in parallel have great potential for the detection of biomarkers and their applications.
NGS in cancer biomarker discovery
Sequencing technologies, particularly NGS, are unbiased, complete and detailed in capturing the heterogeneity of cancer and the rapidly changing genetic landscape in cancer, and can significantly improve the chances of identifying actionable genetic aberrations. In addition to detecting changes in DNA sequences, NGS can also be used for transcriptome analysis and methylation detection. The Cancer Genome Atlas (TCGA) is a tumor genome initiative, launched in 2006 by the National Cancer Institute and the National Human Genome Research Institute, to use high-throughput genomic analysis technologies to help improve understanding of tumors and improve the prevention, diagnosis and treatment of tumors. Currently, TCGA already contains information on sequencing results, transcriptome analysis, copy number variants, DNA methylation and single nucleotide variants, covering 33 tumor types.
5 types of biomarkers that can be identified by NGS
- Genetic variants
Mutations in the form of single nucleotide variants, insertions, deletions and other structural variants are associated with cancer development, progression and metastatic potential as well as the effectiveness of treatment. For example, mutations in BRCA1/2, KRAS, PTEN and others have been identified as prognostic and predictive markers. NGS-based assays can therefore be used to detect known and new cancer mutations. Another advantage of using NGS for biomarker testing is that genetic material from archived samples such as FFPE blocks can also be used.
While genetic aberrations in individual genes can inform drug development and diagnosis, cancer cells often have mutation pathways in multiple genes, making a more detailed mutation landscape of great value. Some NGS Panels designed on the basis of a priori knowledge are now able to detect dozens of cancer-associated genes simultaneously, such as the majority of patients carrying BRCA1/2 mutations. In addition, many of the anti-cancer drugs approved by the FDA in the last decade have targeted specific genetic aberrations. For example, gefitinib has been approved for lung adenocarcinoma carrying an EGFR mutation and virofenib has been approved for melanoma patients carrying a V600E BRAF mutation.
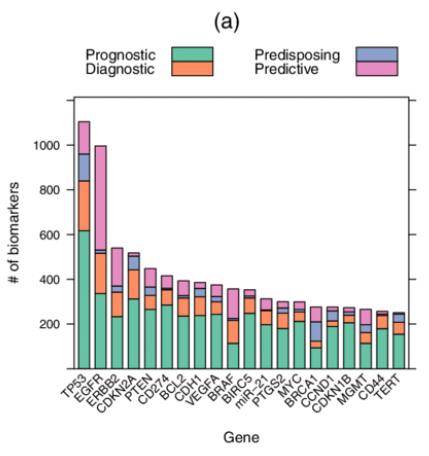 Top genes as cancer biomarker. (Lever J et al., 2019)
Top genes as cancer biomarker. (Lever J et al., 2019)
- Gene expression profiling
There are many isoforms of a gene and a large number of studies have shown that specific gene isoforms are expressed in cancer in an altered manner. Transcriptome analysis using NGS has very distinct advantages as expression data is captured at the isoform level, which can provide unique and important insights into cancer progression and metastasis. Breast cancer, colon cancer and glioblastoma are examples of cancer types where gene expression profiling is widely used. In addition, biomarker analysis by whole blood RNA-Seq allows analysis of expression differences in blood samples, such as the more cutting-edge liquid biopsy, which is a blood test instead of a tumor tissue test, and can provide references and insights for monitoring cancer recurrence and assessing efficacy.
- Epigenetic modifications
Epigenetic modifications are DNA changes that are independent of DNA sequence variation. epigenetic modifications such as DNA methylation, histone acetylation and methylation have profound effects on gene expression and are classical biomarkers that can be used for cancer detection and therapeutic intervention development. Examples of epigenetic modifications used as biomarkers include hypermethylation of GSTP1 in prostate cancer patients and DAPK and hypermethylation of the RASSF1A gene in bladder cancer. High throughput technologies such as epigenome sequencing are well suited to identify epigenetic modifications as biomarkers in cancer.
- MicroRNA
MicroRNAs are small (19-22 nucleotides long) single-stranded RNA molecules that play a key role in the regulation of gene expression by targeting mRNA for degradation or by inhibiting translation. Regulation of gene expression mediated by microRNAs is important in development, differentiation and cell growth, and therefore microRNAs may play a key role in carcinogenesis. miR15 and miR16 have been shown to play an important role in chronic lymphocytic leukemia (CLL). miR206 is downregulated in 93% of breast cancer cases. The role of microRNA in cancer is a very active area of research and it is likely that many key molecules remain to be discovered.
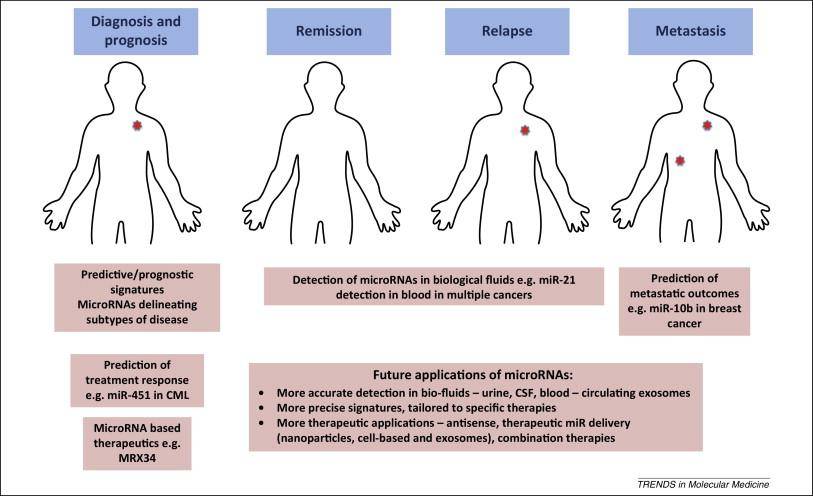 MicroRNAs in cancer (Hayes J et al., 2014)
MicroRNAs in cancer (Hayes J et al., 2014)
- Circulating tumor DNA
Circulating tumor DNA (ctDNA) refers to the release of somatic DNA from tumor cells into the circulatory system, either through shedding or when apoptosis occurs. ctDNA is the DNA fragment from the tumor genome that is constantly flowing in the human circulatory system carrying certain characteristics (including mutations, deletions, insertions, rearrangements, copy number abnormalities, methylation, etc.). Circulating tumor DNA is cfDNA (cell-free DNA) derived from tumor cells and belongs to a type of cfDNA. Currently, multigene or even whole genome analysis of ctDNA samples is commonly performed using NGS, which allows the detection of various mutations including ALK fusions with ROS1 by employing RNA probe capture techniques.
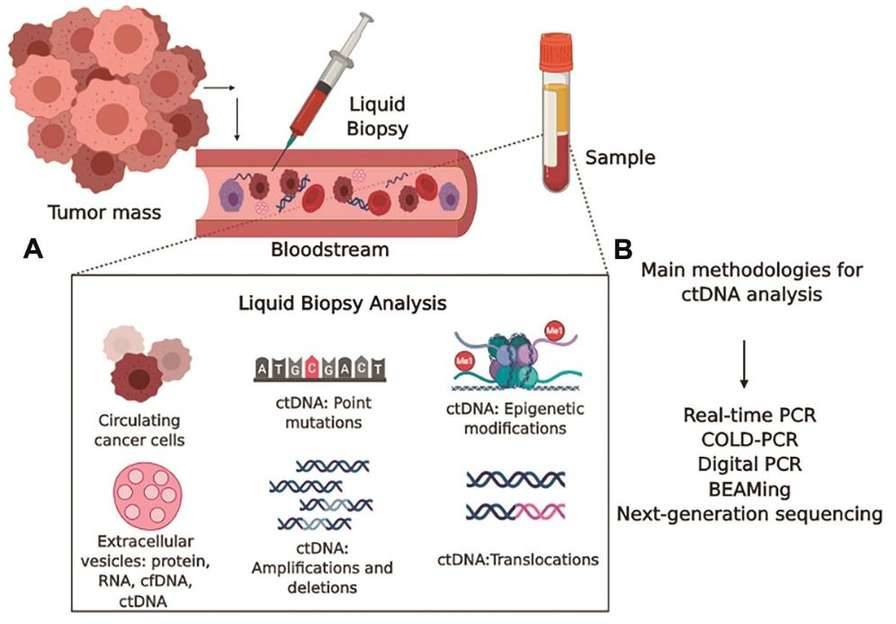 ctDNA as a Cancer Biomarker (Pessoa L S et al., 2020)
ctDNA as a Cancer Biomarker (Pessoa L S et al., 2020)
Challenges of NGS in cancer biomarker discovery
- Variants of Undetermined Significance
Variants of Undetermined Significance (VUS) are DNA variants whose functional impact has not been fully characterized. The presence of VUS in genomic regions important for the disease under consideration can create a great deal of uncertainty in determining future action and can therefore sometimes be counterproductive. Tests for biomarker identification are expected to yield precise and specific information that can be reliably used to guide disease management, and NGS-identified VUS present a significant challenge to their utility as standard tests.
- Complex data analysis and interpretation
In the context of cancer biomarker discovery, NGS generates vast amounts of complex data, including genetic variations, gene expression patterns, and epigenetic modifications. This complexity can make data analysis and interpretation challenging, particularly for researchers without specialized computational and statistical skills. Additionally, the development of robust and reliable bioinformatics tools for NGS data analysis and interpretation is essential. These tools must be able to identify and annotate genetic variants accurately, predict their functional impact, and prioritize those that are most likely to be clinically relevant.
References:
- Desai A N, Jere A. Next-Generation Sequencing for Cancer Biomarker Discovery[J]. Next Generation Sequencing in Cancer Research, Volume 2: From Basepairs to Bedsides, 2015: 103-125.
- Pessoa L S, Heringer M, Ferrer V P. ctDNA as a cancer biomarker: A broad overview[J]. Critical reviews in oncology/hematology, 2020, 155: 103109.
- Lever J, Jones M R, Danos A M, et al. Text-mining clinically relevant cancer biomarkers for curation into the CIViC database[J]. Genome medicine, 2019, 11: 1-16.
- Hayes J, Peruzzi P P, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy[J]. Trends in molecular medicine, 2014, 20(8): 460-469.


 Sample Submission Guidelines
Sample Submission Guidelines
