High-throughput Single-cell V(D)J Sequencing: Introduction, Principle and workflow
Diversity and molecular mechanisms of specific immunity
The immune system is divided into non-specific and specific immunity according to whether it is "targeted" to the "non-self" or not. The molecular basis of specific immunity "targeting" is assumed by the T-cell immune receptor/B-cell immune receptor protein (TCR/BCR) molecules expressed on the surface of T/B lymphocytes, and one "non-self" corresponds to one TCR or BCR.
There are countless pathogens in nature, but human cells have only about 25,000 genes, so how do they meet the need for TCR/BCR diversity? The answer is related to the high degree of rearrangement of the genes encoding TCR/BCR.
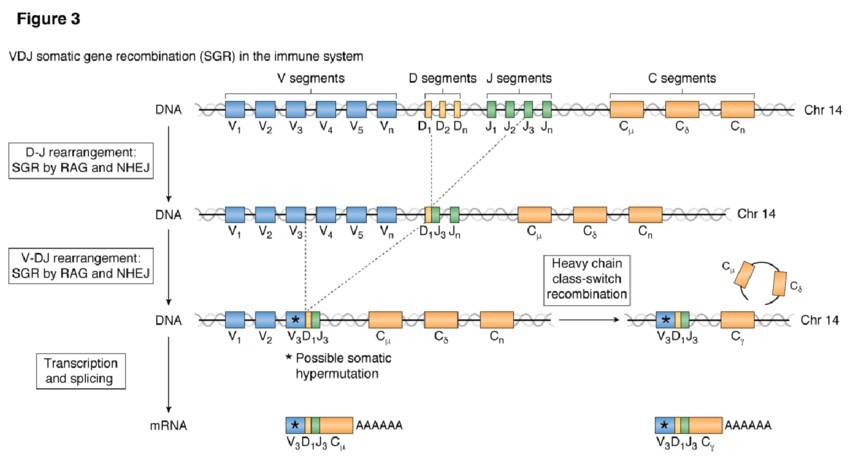 VDJ recombination in the immune system. (Kaeser et al. 2020)
VDJ recombination in the immune system. (Kaeser et al. 2020)
The highly variable regions in the TCR/BCR molecule are encoded by combinations of V, D, and J genes, and a wide variety of TCR/BCRs are generated through highly gene rearrangements such as modular combinations of permutations and mutations. In the BCR, for example, there are about 40 different V modules, 25 different D modules, and 6 different J modules in the human gene. The B cells select one of the V, D, J, and C gene modules, respectively B cells select one of them and splice them together to assemble a mature BCR coding gene, there will be 2400 combinations.
Similarly, T cells undergo gene rearrangement to generate diverse TCRs. However, the process of TCR gene rearrangement involves the selection and rearrangement of variable (V), joining (J), and constant (C) gene segments. The combination of different V and J gene segments contributes to the diversity of TCRs.
Please refer to our article Strategy Overview for Next-Generation Sequencing-based TCR Profiling for detailed information.
Regarding the differences in the response to the new coronavirus (SARS-CoV-2), the variability in TCRs and BCRs among individuals can contribute to variations in immune responses. Some people may have TCRs and BCRs that are better suited for recognizing and combating the virus, resulting in a milder or asymptomatic infection. Conversely, individuals with TCRs and BCRs that are less effective in recognizing the virus may experience more severe illness.
To gain a deeper understanding of the types and quantitative differences of TCR and BCR molecules in immune responses, researchers can employ high-throughput single-cell V(D)J sequencing. This technology allows for the analysis of individual T or B cells, providing insights into the diversity and composition of the immune repertoire at a single-cell level. Such studies can contribute to our understanding of immune responses to infections, autoimmune diseases, and other immune-related disorders.
Introduction of single-cell V(D)J sequencing technology
High-throughput single-cell V(D)J sequencing technology is capable of capturing mRNA from thousands of single cells at a time at the single-cell level, and obtaining V(D)J gene expression information for each T-lymphocyte and B-lymphocyte while obtaining the full gene expression profile of each cell.
To meet the needs of "high-throughput" and "single-cell", high-throughput single-cell V(D)J sequencing is mainly achieved by using droplet microfluidic chips and microspheres with molecular tags.
A single-cell suspension obtained by sample dissociation, molecularly labeled microspheres, reverse transcription kit, and droplet generation oil are added to the spiked wells of the droplet microfluidic chip.
By controlling the flow rate of microspheres, cells, and reagents in the microfluidic flow channel, the microspheres, cells, and reagents can be paired in the desired ratio and finally enter the oil phase through the aqueous phase, which can enclose the cells and microspheres in the oil-in-water, thus forming a single-cell state.
After the cells enter the oil-in-water, the cell membrane ruptures to release mRNA molecules. mRNA molecules are captured by structures such as polyT or C-C-C at the end of the million single-stranded oligos carried on the microspheres. Each oligo carries both a Cell Barcode and a Transcript Label (molecular barcodes), so that even if tens of thousands of cells are mixed for subsequent reverse transcription and library construction experiments, the library data can still be split to each cell and restored to the true gene expression level by using the Cell Barcode and molecular barcodes during the data analysis stage.
Please read our article Immune Repertoire Sequencing: Introduction, Workflow, and Applications for more information.
Workflow of single-cell V(D)J sequencing
High-throughput single-cell V(D)J sequencing also requires the dissociation of tissue samples into single-cell suspensions, followed by the formation of water-in-oil structures with microfluidic chips to complete single-cell separation, followed by nucleic acid capture and transcriptome library and TCR/BCR library construction.
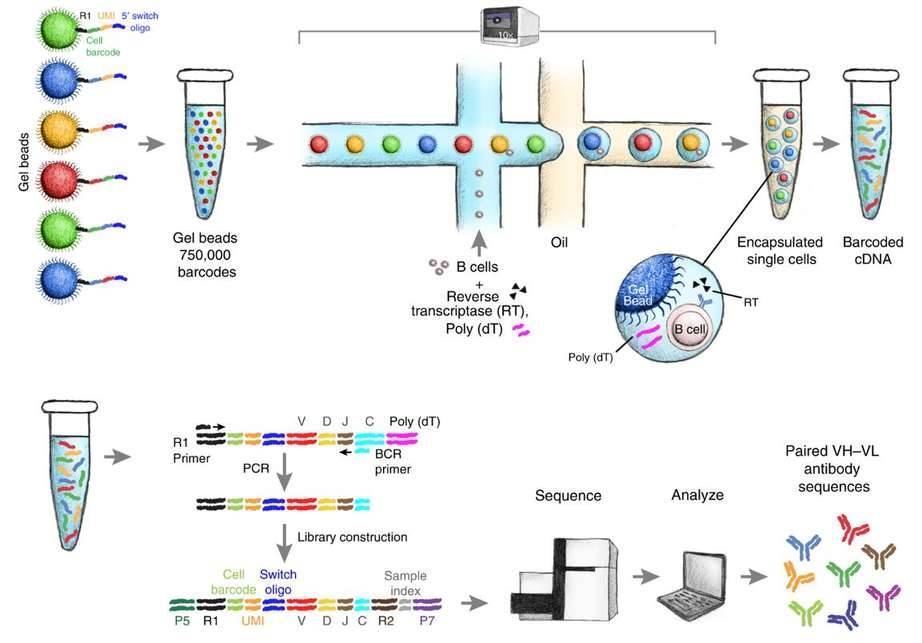 Massively parallel single-cell B-cell receptor sequencing. (Goldstein et al., 2019)
Massively parallel single-cell B-cell receptor sequencing. (Goldstein et al., 2019)
The main steps from cell isolation to library construction are as follows:
Step 1: In the water-in-oil structure, the cell membrane is dissolved to release intracellular mRNA molecules.
Step 2: mRNA molecules are synthesized into single-stranded cDNA by reverse transcriptase. cDNA molecules are captured by oligonucleotides with Cell Barcode, and further synthesized into double-stranded DNA molecules. In this process, each DNA molecule is tagged with a cell label.
Step 3: Take some of these double-stranded cDNA molecules to complete the construction and sequencing of a whole gene expression profile transcriptome library (cDNA molecules from all cells are mixed together in this process to build a multicellular hybrid library).
Step 4: Take some of the double-stranded cDNA molecules and complete the V(D)J library construction and sequencing by combining the primers of the conserved region of V(D)J gene (all the cDNA molecules of the cells are mixed together in this process to build a multicellular hybrid library).
In the subsequent data analysis phase, the transcriptome library and V(D)J library can be reduced to the single-cell level based on cell tags, which can be used to understand the gene expression profile and V(D)J gene expression information of each cell. The full gene expression profile of each cell, combined with Marker genes specifically expressed in different cell types, allows cellular annotation of each cell to understand those types of cells present in each sample, the number of each cell, and the gene expression of each cell. In combination with the V(D)J library it is possible to know which V(D)J gene is expressed in each T or B lymphocyte and which TCR/BCR molecule corresponds to it.
References:
- Kaeser, Gwendolyn, and Jerold Chun. "Brain cell somatic gene recombination and its phylogenetic foundations." Journal of Biological Chemistry 295.36 (2020): 12786-12795.
- Goldstein, Leonard D., et al. "Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies." Communications biology 2.1 (2019): 304.


 Sample Submission Guidelines
Sample Submission Guidelines
