Immune Repertoire Sequencing: Introduction, Workflow, and Applications
Introduction to Immune Repertoire Sequencing
The adaptive immune system plays an extremely important role in counteracting against infectious pathogens and shielding the human body. The foundation of the adaptive immune system is fundamentally reliant upon the enormous diversity of T and B cell receptors (TCRs and BCRs) that can recognize epitopes from an astronomical number of different exogenous antigens and endog-enous host factors. The immune repertoire is known as the collection of functionally diverse B and T-cells in the circulatory system, which is a key measure of immunological complexity.
The study of Immune repertoire is crucial to understand the response of adaptive immunity, and ultimately, sheds light on the discovery of novel infectious agents and holds the invaluable potential to aid in antibody or vaccine development, clinical diagnosis, treatment, and prevention.
The traditional strategies for studying the immune repertoire, which includes spectratyping, Sanger sequencing et al., are laborious, costly, and are insufficient for generating a high-resolution picture of the immune repertoire. NGS offers a powerful approach capable of analyzing the immune repertoire in a high-throughput manner. By providing enormous sequencing data, NGS has created an unprecedentedly high-resolution picture of the immune repertoire, which provides great potential for revealing dynamic changes in clonal populations during antigen stimulation.
Immune Repertoire Sequencing Workflow
The workflow of immune repertoire sequencing is composed of six steps. The first step is cell isolation. This will then be followed by template purification and molecule amplification. The next step would be Ig-seq library construction and sequencing. This will be followed by data processing which involves filtering, alignment, and clustering of data. Lastly, the data will be analyzed considering the repertoire diversity, dynamics, and clonality.
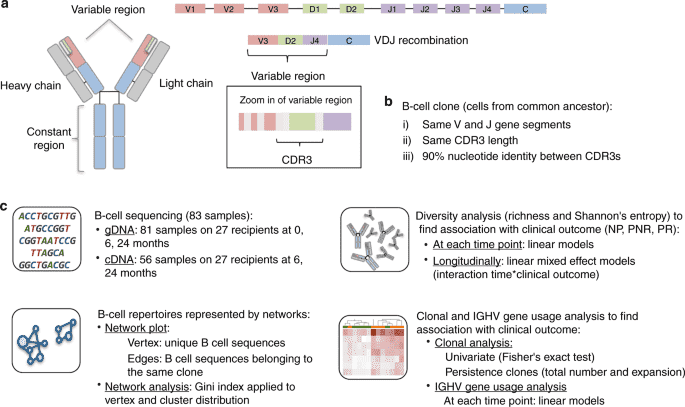
Figure 1. An overall study pipeline of B cell immune repertoire sequencing. (Pineda, 2019)
Applications of Immune Repertoire Sequencing
Immune repertoire sequencing is applied in the following areas: (1) antibody discovery and vaccine development, (2) biomarker discovery, (3) understanding the mechanisms of immune repertoire development, (4) immunodeficiency identification, and (5) a clinical diagnostic tool.
Biomarkers for cancer diagnosis, subtyping, and prognosis
One important field for translational research and medicine of TCR/BCR HTS sequencing is the minimal residual disease (MRD) detection for lymphatic hematological tumors, including T/B cell-derived leukemia and lymphoma. The sensitivity and prognosis use for NGS in comparison with the traditional techniques such as flow cytometry and allelic-specific PCR have been recurrently investigated ever since the beginning of this field.
Comprehensive Analysis of Gut-Microbiota-Reactive Immune Repertoires
Although high-throughput immune cell repertoire analysis in the context of host-microbiome interaction is an area of open research, the most promising application of this approach is comprehensive detection of gut-microbiota-reactive T- or B-cell clonotypes in the circulating blood or peripheral tissues of the hosts. Very importantly, by using live-cell CD154 expression assay, it has recently been shown that healthy human adults possess a substantial amount of circulating CD4+ T cell populations reactive against bacterial lysates of gastrointestinal commensals at frequencies of 40–500 per millions of total CD4+ T cells depending on each bacterial species. The majority of these gut-microbiota-reactive CD4+ T cells had a memory phenotype with relatively high expression of mucosa homing receptors and a Th17 marker (CD161) and was further enriched in the intestinal tissues.
Gut Microbiota as a Possible Origin of TCR Cross-reactivities
T-cell cross-reactivity is an important area of ongoing studies in system immunology and probably one of the not yet fully recognized chief principles of an adaptive immune response. Consistent with this hypothesis and in contrast to the previous general belief, an elegant study using TCRβ transgenic mice proved that the vast majority of negatively selected TCRs are autoreactive and endowed with cross-reactivities against multiple MHC haplotypes, while cross-reactive TCRs are very infrequent among preselection TCRs.
References:
- Pineda S, Sigdel TK, Liberto JM, et al. Characterizing pre-transplant and post-transplant kidney rejection risk by B cell immune repertoire sequencing. Nature communications. 2019, 10(1).
- Liu X, Wu J. History, applications, and challenges of immune repertoire research. Cell biology and toxicology. 2018, 34(6).
- Rubelt F, Busse CE, Bukhari SA, et al. Adaptive immune receptor repertoire community recommendations for sharing immune-repertoire sequencing data. Nature immunology. 2017, 18(12).
- Friedensohn S, Khan TA, Reddy ST. Advanced methodologies in high-throughput sequencing of immune repertoires. Trends in biotechnology. 2017, 35(3).
- Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Current opinion in immunology. 2013, 25(5).


 Sample Submission Guidelines
Sample Submission Guidelines
