Introduction To DNA Molecular Markers
Molecular markers represent a specific category within the broader classification of genetic markers. In a general context, genetic markers encompass traits that enable the tracking of the inheritance of various genetic characteristics present on a chromosome, a particular chromosomal segment, or a specific gene location within a family lineage. Genetic markers are characterized by two fundamental features: heritability and the ability to be distinguished. Essentially, any genetic mutation leading to discernible differences in phenotype within a particular organism can fulfill the role of a genetic marker. This makes genetic markers a vital tool in genetic research and analysis.
What Are Molecular Markers
Genetic markers, which originate from variances in nucleotide sequences among individuals, offer a direct representation of genetic polymorphism at the DNA level. They serve as a direct means to gain insights into distinctions within the genomic DNA of various biological individuals or populations. This fundamental concept underscores their importance in genetic research and analysis.
Advantages of Molecular Markers
- They are presented in DNA form and are not constrained by factors such as tissue, developmental stage, season, or environment. They do not suffer from issues related to expression or lack thereof, exhibiting stability.
- They are numerous, distributed throughout the entire genome.
- They exhibit high polymorphism, with many existing allelic variations in nature.
- Many markers show a co-dominant nature, allowing differentiation between homozygous and heterozygous genotypes, making them convenient for selecting recessive agronomic traits.
- They have a neutral effect, not influencing the expression of the target traits and having no linkage with undesirable characteristics.
- Their detection methods are simple and rapid.
Comparison of Co-Dominant and Dominant Markers
Two important terms in the context of molecular markers are dominant markers and co-dominant markers. In simple terms, molecular markers that can distinguish between heterozygous genotypes are considered co-dominant markers.
| Co-Dominant Marker | Dominant Marker | |
| Definition | A marker in which both alleles from two homozygous parents can be distinguished in heterozygous F1 individuals. It can differentiate between homozygous and heterozygous genotypes. | A genetic marker that can detect only dominant alleles and cannot differentiate between homozygous and heterozygous genotypes. |
| Representation | Polymorphism in the presence and length of amplified fragments. | Presence or absence of amplified fragments. |
Molecular Marker Technologies Applications
Construction of Genetic Linkage Maps: Molecular markers are instrumental in the construction of genetic linkage maps, which provide valuable insights into the location and order of genes on chromosomes.
Gene/QTL Mapping: These markers play a crucial role in the localization of genes and Quantitative Trait Loci (QTLs), aiding in the identification of genetic factors responsible for specific traits.
Molecular Marker-Assisted Breeding: Molecular markers assist in selective breeding by allowing for the targeted transfer of desired traits from one organism to another, enhancing the efficiency of breeding programs.
Analysis of Plant Genetic Diversity: Molecular markers are utilized to assess and characterize genetic diversity within plant populations, aiding in the conservation and utilization of genetic resources.
Variety and Quality Purity Assessment: They are employed to identify and determine the genetic purity and varietal identity of plant materials, ensuring the integrity of crop varieties.
Disease Detection: Molecular markers are employed in the detection of genetic markers associated with diseases, facilitating disease diagnosis and management in both plants and animals.
Types of Molecular Markers
First Generation - RFLP (Restriction Fragment Length Polymorphism)
The application of first-generation RFLP markers has declined over time. RFLP, which stands for Restriction Fragment Length Polymorphism, involves the detection of variations in DNA fragments resulting from changes such as base substitutions, insertions, deletions, or duplications that affect the recognition sites of restriction endonucleases on the plant genome.
The RFLP principle involves digesting the genomic DNA of different individuals using restriction endonucleases, which produces DNA fragments of varying sizes. These fragments are separated through electrophoresis and transferred to a hybridization membrane. A specific DNA fragment, labeled with isotopes or non-isotopes, serves as a probe. The probe hybridizes with the enzyme-digested fragments, revealing differences in length among fragments with homologous sequences to the probe.
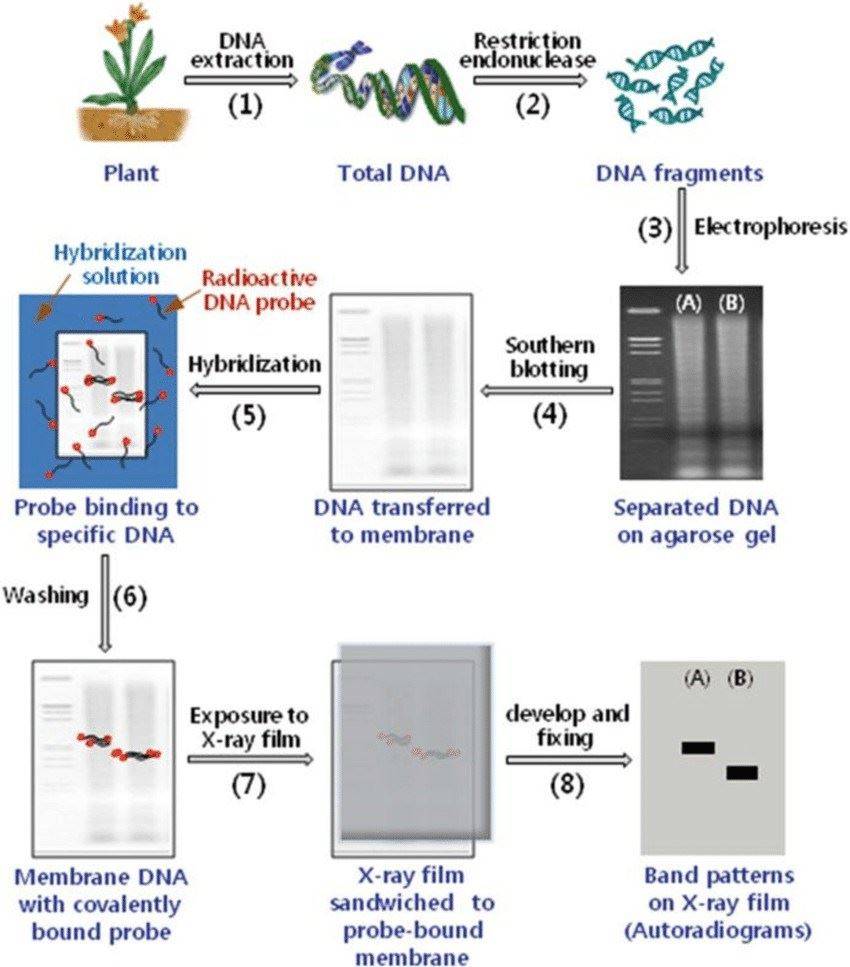 Restriction fragment length polymorphism (RFLP) procedure (Sidharth Mishra et al,. Advances in Veterinary Sciences 2019)
Restriction fragment length polymorphism (RFLP) procedure (Sidharth Mishra et al,. Advances in Veterinary Sciences 2019)
RFLP (Restriction Fragment Length Polymorphism) reflects differences in the lengths of DNA fragments generated after digestion with restriction endonucleases, thereby revealing the distribution of different enzyme recognition sites at the DNA molecular level.
Advantages of RFLP markers include their predominantly co-dominant nature and their high reproducibility and stability.
However, RFLP markers have several drawbacks, including complex experimental procedures, extended detection periods, high costs, and limited suitability for large-scale molecular breeding. Additionally, the use of radioactive isotopes in detection can lead to contamination. Non-radioactive labeling methods tend to be more expensive, resulting in weaker hybridization signals and reduced sensitivity.
PCR-Based Molecular Marker Technologies
The second-generation PCR-based molecular marker technologies are currently the most widely utilized in molecular genetics. Polymorphic markers detected through PCR, which encompasses both direct PCR and PCR combined with enzymatic digestion approaches, fall under the category of PCR markers. These markers include RAPD, SSR, STS, CAPS, and dCAPS, among others.
RAPD, Random Amplified Polymorphism DNA, involves the use of short, random primers typically spanning 8-10 base pairs to amplify genomic DNA through PCR. When the distance between binding sites for two reverse complementary primers meets the conditions for PCR amplification, an amplification product is generated. Any insertions, deletions, or mutations occurring between primer binding sites or at the binding sites result in the presence or absence of bands in the electrophoretic pattern of PCR products, giving rise to RAPD markers.
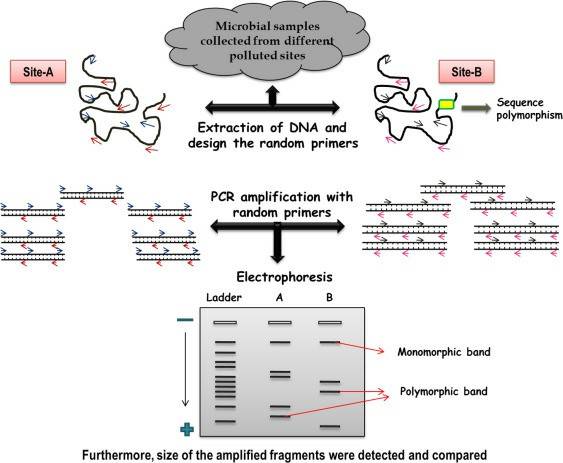 Schematic illustration of random amplified length polymorphism analysis. (Satyanarayan Panigrahi et al,. Microbial Diversity in the Genomic Era 2019)
Schematic illustration of random amplified length polymorphism analysis. (Satyanarayan Panigrahi et al,. Microbial Diversity in the Genomic Era 2019)
Benefits of RAPD Markers:
A straightforward method with swift detection.
Minimal DNA quantity is needed, and it has lenient DNA quality prerequisites.
The same set of primers can be applied for genomic analysis across diverse organisms, facilitating the scrutiny of complete genomes.
Limitations of RAPD Markers:
They are dominant markers and cannot distinguish between dominant homozygous and heterozygous genotypes, leading to incomplete data.
The utilization of short primers in RAPD markers renders the PCR process susceptible to experimental variables, resulting in the presence of multiple bands and diminished result consistency.
SSR markers, or Simple Sequence Repeat markers, also known as microsatellite DNA or short tandem repeats (STRs), represent a class of DNA sequences composed of a repetitive unit of several (1-6) nucleotides. Common repeat motifs include (TG)n, (GA)n, (AAT)n, or (GACA)n, with (AT)n being the most prevalent in plant genomes. These core sequences are widely distributed in eukaryotic and some prokaryotic genomes, found randomly in nuclear DNA, chloroplast DNA, and mitochondrial DNA. SSRs typically have lengths of 100 base pairs or less.
The principle behind SSRs lies in the high conservation of single-copy sequences on both sides of the microsatellite DNA. Thus, primers can be designed based on these flanking sequences for PCR amplification, and the size of PCR products can be determined through electrophoresis. The high polymorphism of SSR markers primarily results from variations in the number of tandem repeats within the core sequence.
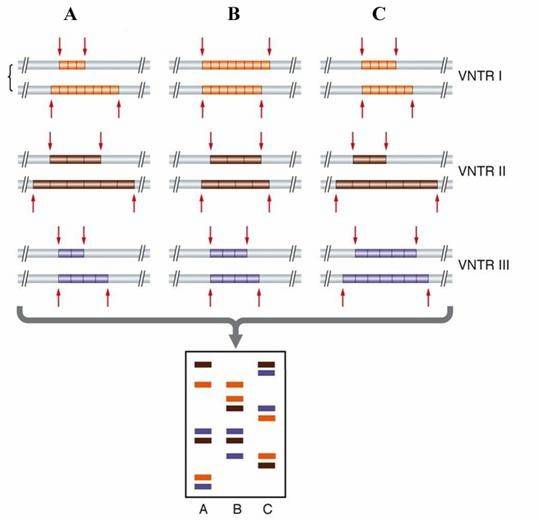 Principle of the SSR marker. (Ksenija Taski-Ajdukovic Research Signpost 2015)
Principle of the SSR marker. (Ksenija Taski-Ajdukovic Research Signpost 2015)
Advantages of SSR Markers:
Represent co-dominant markers, allowing differentiation between heterozygotes and homozygotes.
Require small DNA sample quantities and are not overly demanding in terms of DNA quality.
Exhibit good repeatability and high reliability.
Display a high degree of allelic diversity, resulting in significant polymorphism.
Limitations of SSR Markers:
Knowledge of the DNA sequence at both ends of the repeat motif is essential. In cases where this information is not accessible in DNA databases, sequencing and primer design become necessary, resulting in increased developmental expenses.
You may interested in
AFLP Markers, or Amplified Fragment Length Polymorphism, refers to polymorphic markers created through the amplification of restriction enzyme-digested DNA fragments. Differences arise due to single nucleotide mutations, insertions, deletions, and other mutations that lead to changes in restriction enzyme recognition sites. These differences can be detected through selective PCR amplification. AFLP is an evolution of RFLP, with the distinction lying in the detection method. AFLP reveals polymorphisms at restriction enzyme sites and subsequent selective base variations.
The principle behind AFLP is rooted in both restriction enzyme digestion and PCR technology. Genomic DNA is first digested with two restriction enzymes, resulting in DNA fragments of varying sizes. Artificial adapters are then ligated to the ends of these restriction fragments, serving as templates for the amplification reaction. Pre-amplification is performed using the complementary strand of the artificial adapter as a primer. Subsequently, 1-3 selective bases are added to the pre-amplified products as primers for selective amplification. Polyacrylamide gel electrophoresis is used for separation, and polymorphisms are detected based on differences in amplified fragment lengths.
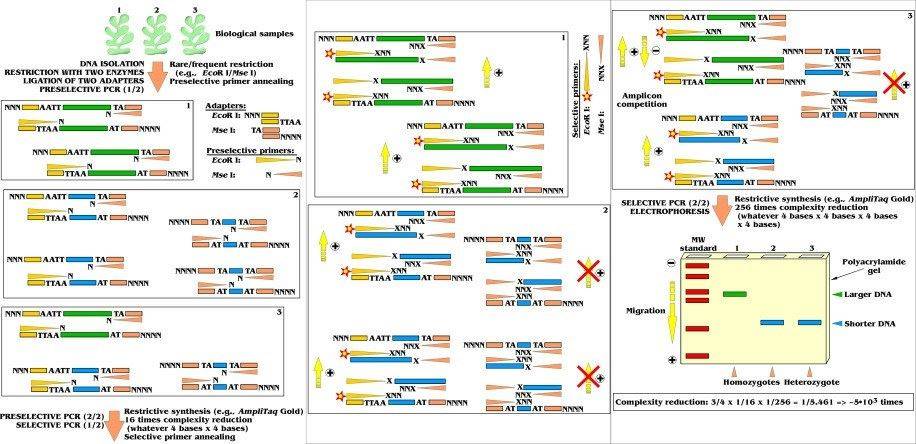 Amplified fragment length polymorphism(G. Dorado, Reference Module in Biomedical Sciences, 2017)
Amplified fragment length polymorphism(G. Dorado, Reference Module in Biomedical Sciences, 2017)
Advantages of AFLP Markers:
Combines the reliability of RFLP technology and the efficiency of PCR technology, enabling the detection of a large number of restriction enzyme-digested fragments in a single reaction. It yields information on 50 to 100 spectral bands in a single run.
Offers dominant (presence or absence of bands) and co-dominant (length differences due to insertions or deletions) markers.
Does not require the development cost of labeled markers but necessitates optimization of specific primers and primer combinations.
Does not require genomic DNA sequence information.
Requires only a minimal amount of DNA.
Disadvantages of AFLP Markers:
High DNA sample quality requirements.
Often employs silver staining, which can be relatively time-consuming and costly.
AFLP markers are not uniformly distributed across chromosomes and tend to cluster in regions near centromeres.
SNP Markers
SNP, which stands for Single Nucleotide Polymorphism, is a third-generation marker that pertains to nucleotide diversity at the genomic level. It arises from variations in a single nucleotide, such as transitions, transversions, insertions, deletions, among others, within a DNA sequence.
SNPs can be detected and analyzed using various techniques. When a known DNA sequence is available, it can be compared to DNA sequence databases, including reference genomes or genomes of closely related species, to identify existing SNPs. Additionally, if SNPs are located at certain restriction enzyme recognition sites, primers can be designed based on the sequences flanking the SNP for PCR amplification. The PCR products can be digested with restriction enzymes and subjected to electrophoresis to transform them into co-dominant CAPS markers. Alternatively, primers can be designed to place the SNP at the 3' end, and the presence or absence of amplified bands is used to convert them into dominant PCR markers. More direct methods include DNA sequencing or using DNA chip hybridization techniques for detection.
You may interested in
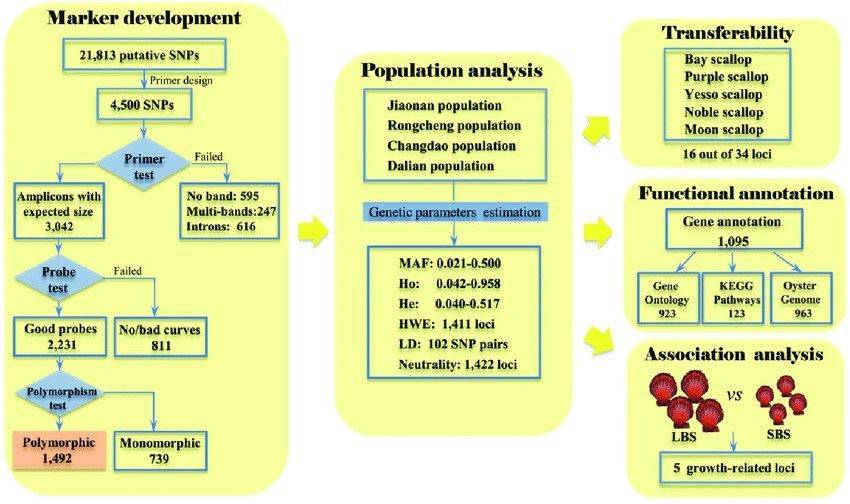 A schematic workflow describing SNP marker development and evaluation
A schematic workflow describing SNP marker development and evaluation
SNP Markers:
Advantages:
Abundant in number and widely distributed in genomes.
High stability.
Co-dominant nature.
Suitable for rapid and large-scale screening.
Disadvantages:
High cost when using sequencing or DNA chip hybridization methods.
Comparison of Various DNA Molecular Markers
| Marker Type | RFLP | RAPD | SSR | AFLP | SNP |
| Genetic Characteristics | Codominant | Mostly Dominant | Codominant | Dominant / Codominant | Codominant |
| Primer/Probe Type | Genomic DNA/DNA-specific, Low Copy Probes | 9-10bp Random Primers | 14-16bp Specific Primers | 16-20bp Specific Primers | Specific Primers |
| Polymorphism Level | Moderate | High | High | High | High |
| Reproducibility/Reliability | High | Low | High | High | High |


 Sample Submission Guidelines
Sample Submission Guidelines
