Comprehensive Overview of m6A Detection Methods
As our comprehension of RNA epigenetics progresses, the significance of accurately discerning N6-methyladenosine (m6A) modifications within RNA becomes increasingly salient. m6A, the predominant internal modification found in eukaryotic messenger RNA (mRNA), orchestrates pivotal roles in diverse biological processes, spanning mRNA stability, splicing, translation, and cellular differentiation. Thus, the imperative to cultivate robust and precise methodologies for m6A detection emerges as paramount in propelling our understanding of RNA epigenetics and its ramifications in health and disease. Within this exhaustive review, we embark on an exploration of various m6A detection modalities, elucidating their underlying principles, merits, and constraints, with particular attention to recent strides within the field.
You may interested in
Antibody-Based Methods
Early techniques like m6A Immunoprecipitation (m6A-IP) hinge on antibodies to profile RNA m6A modifications. Such methods capitalize on antibody selectivity to enrich m6A-modified RNA from total samples. Yet, these methods bear limitations. Antibody specificity and nonspecific binding may yield false positives, impinging on detection precision. Furthermore, antibody reliance can introduce batch variability, potentially impeding reproducibility.
Chemical-Based Methods
Chemical-based methodologies present viable alternatives for m6A detection, circumventing the reliance on antibodies. Notable among these is m6A-selective chemical labeling (m6A-SEAL), employing specific chemical probes to tag m6A sites within RNA. Subsequent sequencing facilitates precise nucleotide-level identification of m6A modifications. Similarly, miCLIP (m6A individual-nucleotide resolution UV crosslinking and immunoprecipitation) merges UV crosslinking, immunoprecipitation, and chemical modification, affording high-resolution mapping of m6A sites. Chemical approaches confer benefits such as diminished antibody dependency and enhanced resolution. Nonetheless, their adoption may be hindered by intricate experimental protocols and susceptibility to background noise, thus limiting widespread application.
Enzyme-Based Methods
Enzyme-based methodologies emerge as promising avenues for m6A detection, harnessing engineered enzyme catalysis to modify m6A-laden RNA molecules. An exemplar in this realm is DART-seq (Deamination of Adenosines, RNA Targeting and Sequencing), employing fusion proteins housing tailored domains for targeted RNA modification. The DART fusion protein, integrating an RNA-binding domain with an editing domain, affords precise identification of m6A sites at single-nucleotide resolution. Recent strides in DART-seq, including enhanced fusion proteins with augmented m6A recognition, amplify its efficacy for m6A mapping. Enzyme-driven approaches proffer heightened specificity and sensitivity vis-à-vis antibody-based modalities, rendering them indispensable in RNA epigenetics investigations.
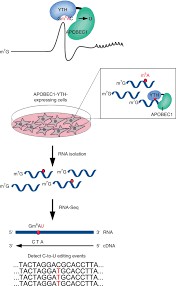 DART-seq
DART-seq
Sequencing-Based Methods
High-throughput sequencing methodologies, exemplified by MeRIP-seq (Methylated RNA Immunoprecipitation Sequencing) and Nanopore RNA Sequencing, are pivotal in elucidating m6A modifications across the transcriptome. Leveraging these approaches, researchers gain profound insights into the distribution and dynamics of m6A modifications within RNA molecules.
MeRIP-seq: Unraveling m6A Distribution with Antibody Immunoprecipitation
MeRIP-seq capitalizes on antibody affinity for m6A to selectively enrich m6A-modified RNA fragments from total RNA samples. This method provides researchers with a means to map m6A sites across the transcriptome with considerable resolution. By precipitating m6A-modified RNA fragments, MeRIP-seq affords a panoramic view of m6A distribution, facilitating the discernment of m6A-modified regions and their functional relevance. However, MeRIP-seq may encounter challenges such as antibody specificity and nonspecific binding, potentially affecting the precision of m6A detection.
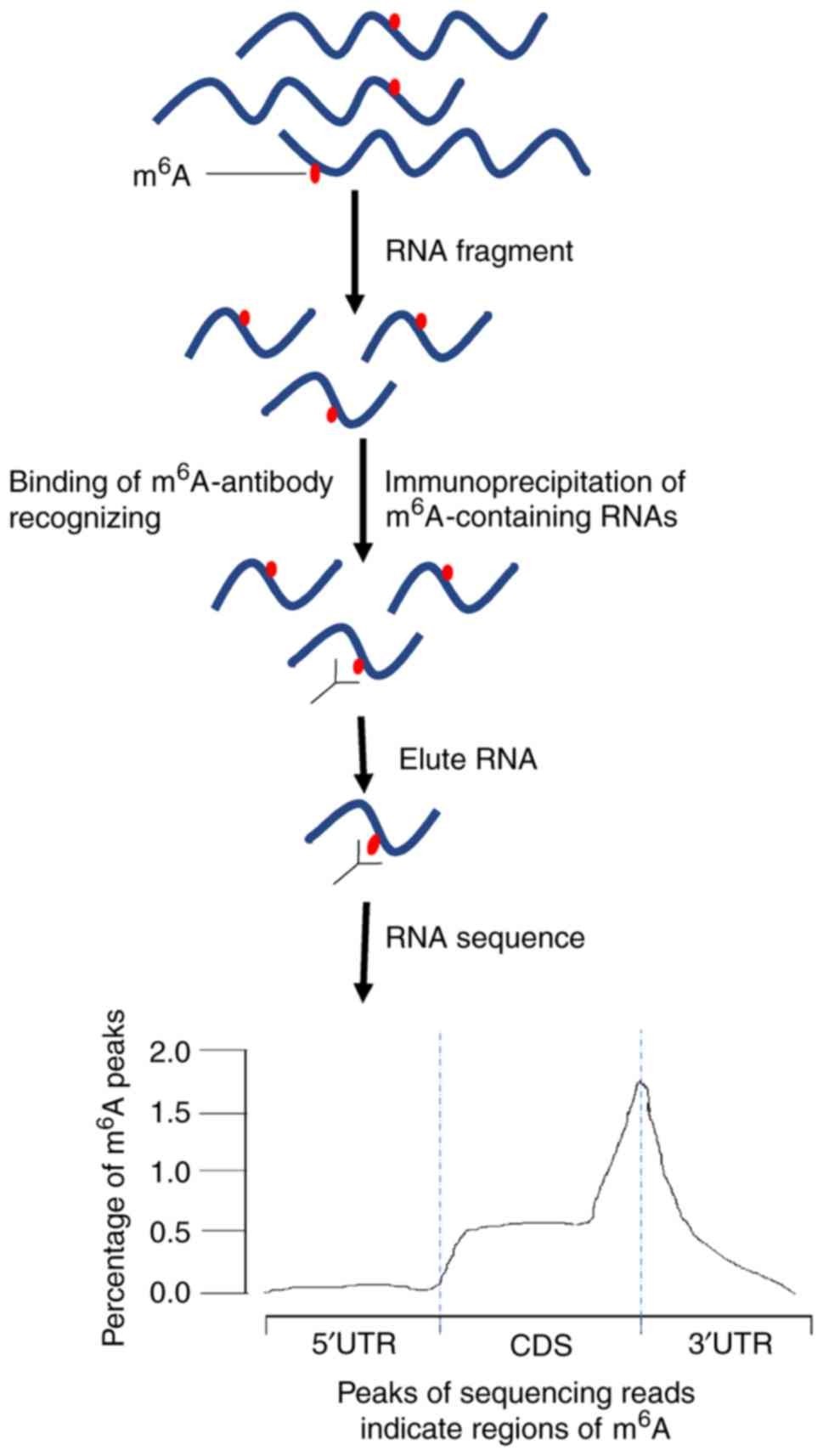 MeRIP-seq
MeRIP-seq
Nanopore RNA Sequencing: Direct Sequencing of RNA Molecules for m6A Detection
Nanopore RNA Sequencing stands as a paradigm shift in m6A detection, facilitating direct analysis of RNA molecules for m6A modifications sans prior enrichment or labeling. This technology harnesses nanopores within a membrane to detect alterations in electrical current as RNA traverses through. The distinct electrical signatures associated with m6A modifications afford precise identification of m6A-modified nucleotides within RNA sequences. Nanopore RNA Sequencing boasts merits including streamlined experimental procedures and real-time capture of dynamic m6A modifications. Nonetheless, addressing challenges like signal noise and data analysis constitutes ongoing areas of research and development.
For more information, refer to "Comparison of M6A Sequencing Methods"
Mass Spectrometry-Based Methods
Mass spectrometry-based methodologies, exemplified by m6A-LAIC-seq (m6A-Level and Isoform Characterization sequencing), present a complementary avenue for m6A detection. These techniques entail direct RNA molecule analysis for m6A presence utilizing mass spectrometry. In m6A-LAIC-seq, RNA molecules undergo initial enzymatic digestion into nucleosides, followed by liquid chromatography coupled with mass spectrometry (LC-MS) for m6A level quantification. While furnishing quantitative data on m6A abundance, mass spectrometry-based approaches are constrained in pinpointing the precise m6A modification locations within RNA sequences.
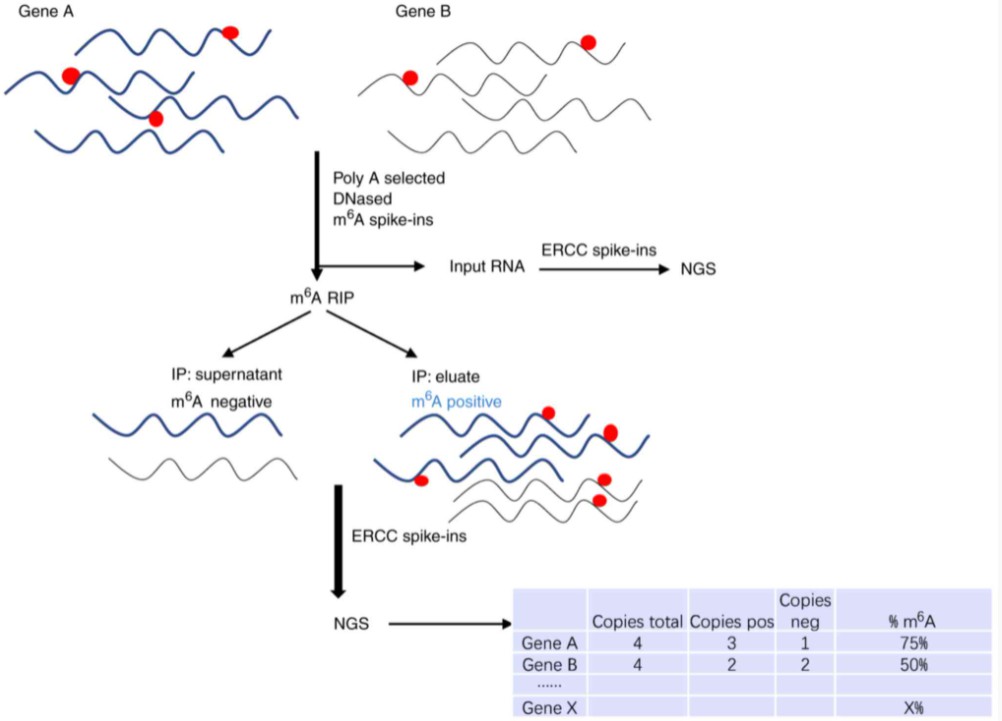 m6A-LAIC-seq
m6A-LAIC-seq
Hybrid Methods
Hybrid methodologies amalgamate diverse detection strategies to surmount the constraints of individual approaches, fostering comprehensive m6A mapping. For instance, m6A-REF-seq (m6A-RECombination and Fragmentation sequencing) merges antibody-based immunoprecipitation with enzymatic fragmentation and sequencing, pinpointing m6A sites with single-nucleotide precision. Likewise, m6A-REF-seq2 combines chemical labeling with immunoprecipitation and sequencing to enhance m6A detection accuracy. Hybrid methods afford benefits including heightened sensitivity, specificity, and resolution vis-à-vis single-method modalities, albeit at the potential cost of increased experimental intricacy and data analysis complexities.
Bioinformatic Methods
Bioinformatic methodologies play a pivotal role in scrutinizing data arising from m6A detection experiments and discerning m6A-modified sites within RNA sequences. These approaches entail the formulation of computational algorithms and software tools to process raw sequencing data, align reads to reference genomes or transcriptomics, and delineate m6A peaks based on enrichment patterns. Prominent bioinformatic tools for m6A analysis encompass HOMER, MACS, and MeTDiff. Leveraging diverse statistical models and machine learning algorithms, these tools adeptly identify m6A-modified regions and predict potential m6A sites within RNA sequences with precision. Bioinformatic methodologies stand as indispensable assets for deciphering experimental findings and unraveling the functional implications of m6A modifications in gene regulation and cellular processes.
Comparison of m6A Detection Methods
| Method | Principle | Characteristics | Advantages | Disadvantages |
| Antibody-Based | Relies on antibodies to enrich m6A-modified RNA fragments from total RNA samples | - Utilizes specific antibodies targeting m6A modifications - Immunoprecipitation-based enrichment of m6A-modified RNA fragments from total RNA samples | - Rapid identification of m6A modifications at the transcriptome level - Can provide quantitative information about m6A abundance | - Selection of antibodies is critical and may lead to false positives or negatives - Batch-to-batch variability in antibody quality can impact reproducibility |
| Chemical-Based | Utilizes specific chemical probes to label m6A sites in RNA | - Chemical labeling of m6A sites in RNA molecules using chemical probes - Enables high-resolution mapping of m6A modifications | - Independent of antibodies, reducing variability in reproducibility - Provides higher resolution and sensitivity compared to antibody-based methods | - Complex experimental procedures may require specialized expertise - Background noise from chemical reactions may affect data accuracy |
| Enzyme-Based | Leverages the catalytic activity of engineered enzymes to modify m6A-modified RNA molecules | - Fusion proteins containing engineered domains for targeted RNA modification - Enables single-nucleotide resolution m6A mapping | - Increased specificity and sensitivity compared to antibody-based methods - Provides single-nucleotide resolution for precise m6A mapping | - Development and optimization of engineered enzymes may be challenging - Requires validation and optimization for each specific application |
| Sequencing-Based | Utilizes high-throughput sequencing technologies to profile m6A modifications | - Relies on high-throughput sequencing platforms to profile m6A modifications across the transcriptome - Provides genome-wide m6A maps and insights into m6A distribution and dynamics | - Allows identification of m6A modifications at a large scale - Offers insights into m6A distribution and dynamics | - Varying resolution and specificity among different sequencing-based methods - Experimental complexity may differ, requiring careful consideration and optimization |
| Mass Spectrometry-Based | Directly analyzes RNA molecules for the presence of m6A modifications using mass spectrometry | - Direct analysis of RNA molecules for the presence of m6A modifications using mass spectrometry - Provides quantitative information about m6A abundance | - Quantitative assessment of m6A abundance and relative levels - Complementary to sequencing-based methods for validation and confirmation | - Limited ability to pinpoint specific m6A modification sites within RNA sequences - Requires specialized equipment and expertise for mass spectrometry analysis |
| Hybrid | Integrates different detection strategies to overcome limitations | - Combination of multiple methods to enhance sensitivity, specificity, and resolution - Offers complementary strengths of individual methods | - Enhanced sensitivity, specificity, and resolution compared to single-method approaches - Overcomes limitations of individual methods | - Complex experimental procedures may require additional optimization and validation - Data integration and analysis may be challenging due to the hybrid nature of the approach |
| Bioinformatic | Analyzes data from m6A detection experiments using computational algorithms | - Utilizes computational algorithms and tools for processing and analyzing raw sequencing data - Identifies m6A-modified sites within RNA sequences based on enrichment patterns | - Essential for interpreting experimental results and gaining insights into functional roles of m6A modifications - Facilitates data interpretation and integration | - Reliance on computational methods may introduce biases and errors - Requires expertise in bioinformatics for accurate data analysis and interpretation |
This detailed table provides a comprehensive comparison of various m6A detection methods, highlighting their underlying principles, characteristics, advantages, and disadvantages with a more specialized focus.
Summary
In summary, the realm of m6A detection methods is multifaceted, each approach bearing distinct advantages and challenges. Antibody-based methods swiftly enable transcriptome-wide m6A identification but may encounter antibody specificity concerns. Chemical-based techniques offer heightened resolution and sensitivity yet demand meticulous optimization and may contend with background noise. Enzyme-driven methodologies afford heightened specificity and sensitivity but mandate rigorous validation per application. Sequencing-based modalities furnish genome-wide m6A maps albeit varying in resolution and experimental complexity. Mass spectrometry-based approaches deliver quantitative m6A abundance assessment yet fall short in pinpointing specific modification sites. Hybrid strategies amalgamate diverse tactics to surmount limitations albeit requiring additional optimization. Bioinformatic methodologies are imperative for data analysis and interpretation, necessitating proficiency in computational biology. Researchers must meticulously assess these aspects to select the optimal method for their research objectives. As m6A detection advances, CD Genomics steadfastly delivers comprehensive solutions to propel RNA epigenetics research forward.
References:
- Capitanchik, C., Toolan-Kerr, P., Luscombe, N. M., & Ule, J. How Do You Identify m6A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets. Frontiers in Genetics, (2020). 11, 398.
- Fan, C., Ma, Y., Chen, S., Zhou, Q., Jiang, H., & Zhang, J. Comprehensive Analysis of the Transcriptome-Wide m6A Methylation Modification Difference in Liver Fibrosis Mice by High-Throughput m6A Sequencing. Frontiers in Cell and Developmental Biology, (2021). 9, 767051.
- Zhu, H., Yin, X., Holley, C. L., & Meyer, K. D. Improved Methods for Deamination-Based m6A Detection. Frontiers in Cell and Developmental Biology, (2022). 10, 888279.
- Liu, H., Begik, O., Lucas, M.C. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10, 4079 (2019).
- Hendra, C., Pratanwanich, P.N., Wan, Y.K. et al. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods 19, 1590–1598 (2022).
- Meyer, K.D. DART-seq: an antibody-free method for global m6A detection. Nat Methods 16, 1275–1280 (2019)
- Zhu W, Wang JZ, Xu Z, Cao M, Hu Q, Pan C, Guo M, Wei JF, Yang H. Detection of N6 methyladenosine modification residues (Review). Int J Mol Med. 2019


 Sample Submission Guidelines
Sample Submission Guidelines
