Comparison of m6A Sequencing Methods
In the domain of epigenetics, the advent of N6-methyladenosine (m6A) modification has emerged as a pivotal regulatory mechanism governing RNA function, with implications spanning gene expression modulation, cellular dynamics, and disease etiology. Amidst the array of techniques employed for m6A detection, m6A sequencing (m6A-seq) has garnered considerable acclaim for its capacity to delineate m6A modifications across the transcriptome with high resolution. In this discourse, we endeavor to furnish a comprehensive exegesis of m6A-seq, elucidating its underlying principles, methodological intricacies, and comparative analyses vis-à-vis alternative m6A detection methodologies.
What is m6A-seq?
m6A-seq stands as a robust high-throughput sequencing methodology meticulously crafted to chart the landscape of m6A modifications pervading the transcriptome. This cutting-edge technique empowers researchers to discern m6A sites with unparalleled single-nucleotide precision, thereby unraveling the intricate distribution and quantitative abundance of m6A modifications embedded within RNA moieties. Leveraging immunoprecipitation techniques coupled with m6A-specific antibodies, followed by rigorous sequencing protocols, m6A-seq emerges as an indispensable tool for delineating the dynamic m6A landscapes across diverse biological milieus, spanning the realms of physiological homeostasis to pathological perturbations engendered by disease states.
You may interested in
M6A Sequencing Principle
At the core of m6A-seq lies the fundamental concept of selectively enriching m6A-modified RNA fragments, coupled with high-throughput sequencing methodologies aimed at discerning and quantifying these pivotal m6A sites. The procedural workflow commences with the fragmentation of RNA molecules, succeeded by immunoprecipitation utilizing m6A-specific antibodies to isolate the m6A-modified RNA fragments. Subsequent sequencing of the immunoprecipitated RNA fragments facilitates the meticulous mapping of m6A sites dispersed across the transcriptome. A judicious application of bioinformatic algorithms then comes into play, facilitating the identification of enriched regions and the elucidation of intricate m6A distribution patterns embedded within the transcriptomic landscape.
MeRIP-Seq: A Key Method in m6A-seq
Methylated RNA immunoprecipitation sequencing (MeRIP-Seq) has emerged as a cornerstone method in the field of m6A-seq, enabling the precise mapping of m6A modifications across the transcriptome. This technique relies on the selective immunoprecipitation of m6A-modified RNA fragments using specific antibodies, followed by high-throughput sequencing to identify enriched regions.
MeRIP-Seq Principle and Application
At the crux of MeRIP-Seq lies the foundational concept of selectively enriching m6A-modified RNA fragments via immunoprecipitation utilizing m6A-specific antibodies. An exemplary investigation conducted by Dominissini et al. (2012) underscored the efficacy of MeRIP-Seq in elucidating the landscape of m6A modifications within the transcriptome of mouse embryonic stem cells (mESCs). Through an integrative approach marrying MeRIP-Seq data with transcriptome-wide RNA sequencing (RNA-seq), the authors unveiled a myriad of m6A-modified sites dispersed across mESC transcripts, thereby unveiling the pervasive presence of m6A modification within both coding and noncoding genomic regions.
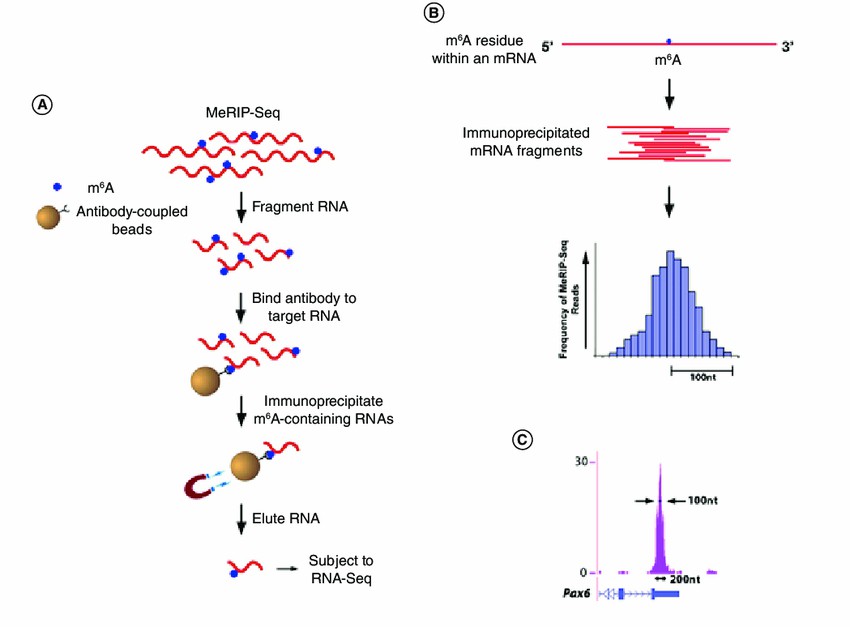 MeRIP-Seq protocol (Joseph Martin 2018)
MeRIP-Seq protocol (Joseph Martin 2018)
MeRIP-Seq Insights into m6A Regulation
MeRIP-Seq has emerged as a potent tool in unraveling the regulatory influence of m6A modifications on gene expression dynamics and cellular phenotypes. In a seminal investigation by Liu et al. (2015), MeRIP-Seq was leveraged to dissect the temporal dynamics of m6A modifications throughout the trajectory of mouse brain development. This insightful study delineated stage-specific patterns of m6A modification within mRNAs intricately linked with neurodevelopmental processes. By shedding light on the nuanced regulation orchestrated by m6A modifications, the findings underscore the pivotal role of m6A-mediated mechanisms in sculpting the landscape of brain development and functional maturation.
MeRIP-Seq Application in Disease Research
Within the landscape of disease investigation, MeRIP-Seq emerges as a pivotal asset in unraveling the intricate involvement of m6A modifications in the pathogenesis of cancer. A seminal inquiry conducted by Li et al. (2017) harnessed MeRIP-Seq to delineate the m6A modification profiles within hepatocellular carcinoma (HCC) cells. Through meticulous analysis, the authors unveiled a cadre of dysregulated m6A-modified transcripts intricately linked with the progression of HCC, thus positing novel candidates for biomarker exploration and therapeutic intervention in combating this formidable malignancy.
Advancements in MeRIP-Seq Technology
Recent advancements in MeRIP-Seq technology have propelled its efficacy to new heights within the realm of m6A research. A pioneering investigation by Ke et al. (2021) unveiled a refined MeRIP-Seq protocol, denoted as m6A-LAIC-seq, which seamlessly integrates MeRIP-Seq with long-read sequencing technology. This innovative amalgamation engenders the detection of m6A modifications at unparalleled single-nucleotide resolution, coupled with the advantageous long-read capabilities, thereby affording a panoramic vista of m6A landscapes embedded within the transcriptome.
m6A Nanopore Sequencing: Advancing the Field of m6A-seq
Recent strides in sequencing methodologies have catalyzed the emergence of nanopore sequencing-based techniques for the detection of m6A modifications. Nanopore sequencing confers a multitude of advantages, notably encompassing long-read capabilities and real-time sequencing, thus positioning it as a promising avenue for m6A-seq endeavors. By directly interrogating RNA molecules as they traverse through nanopores, this technology facilitates the discernment of m6A modifications with exceptional accuracy and resolution. Such advancements hold profound promise for enriching our comprehension of m6A biology and unraveling its intricate implications in health and disease.
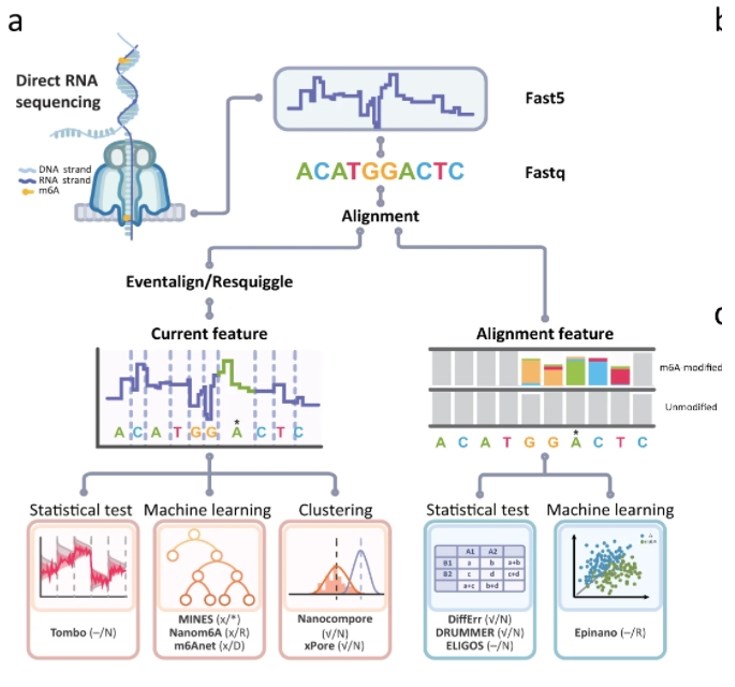 m6A mapping based on the ONT DRS platform (Zhong, ZD., et al,. Nat Commun (2023).
m6A mapping based on the ONT DRS platform (Zhong, ZD., et al,. Nat Commun (2023).
Nanopore Sequencing Principle
Nanopore sequencing has emerged as a groundbreaking modality within the field of genomics, affording the capability to directly sequence RNA molecules in real-time as they traverse through nanoscale apertures. At its essence, this technology hinges upon the detection of alterations in ionic current as RNA molecules transit the nanopore, thereby enabling base identification and sequencing devoid of the requirement for PCR amplification or fluorescent labeling.
Advantages of Nanopore Sequencing
An instrumental attribute of Nanopore sequencing lies in its capacity to furnish long-read sequencing datasets, a facet particularly advantageous for scrutinizing RNA modifications such as m6A. A seminal investigation by Garalde et al. (2018) exemplified the prowess of nanopore sequencing in generating long-read RNA sequencing data boasting single-nucleotide resolution. The authors showcased the ability of nanopore sequencing to accurately discern RNA modifications, including m6A, within lengthy RNA transcripts.
Nanopore Sequencing Application in m6A-seq
Nanopore sequencing represents a groundbreaking advancement in the realm of m6A-seq, facilitating the direct detection of m6A modifications within RNA molecules devoid of chemical interventions or immunoprecipitation procedures. Workman et al. (2019) conducted a pivotal investigation utilizing nanopore sequencing to delineate m6A modifications in human embryonic stem cells (hESCs) with remarkable precision and accuracy. Their study underscored the capability of nanopore sequencing to pinpoint m6A modifications at the level of individual RNA molecules, thereby illuminating the spatial and temporal dynamics governing m6A distribution throughout the transcriptome.
Single-Molecule Resolution
The exquisite resolution afforded by Nanopore sequencing facilitates the comprehensive characterization of m6A modifications at the level of individual RNA molecules, thereby offering unparalleled insights into the heterogeneity and dynamics of m6A. In a seminal investigation, Smith et al. (2020) harnessed the power of nanopore sequencing to explore m6A modifications within the transcriptome of Arabidopsis thaliana. Their study unveiled dynamic fluctuations in m6A modification profiles throughout various developmental stages, thereby shedding light on the pivotal role of m6A in orchestrating the intricate processes underlying plant growth and development.
Future Directions
The burgeoning technology of Nanopore sequencing stands as a pivotal tool poised to propel our comprehension of m6A biology, illuminating its multifaceted implications in health and disease. Future investigations are poised to refine nanopore sequencing protocols tailored specifically for m6A detection, enhancing the precision of base-calling processes, and seamlessly integrating nanopore sequencing datasets with complementary omics technologies to facilitate comprehensive m6A-seq analyses. With ongoing strides in nanopore sequencing methodologies, the landscape of m6A-seq research is primed for continued breakthroughs and transformative discoveries.
Comparison of m6A-seq Methods
MeRIP-Seq: A Classic Approach
MeRIP-Seq stands as a cornerstone technique in the study of m6A modifications within RNA, offering a suite of advantages such as heightened sensitivity and specificity in m6A detection, compatibility with scant RNA inputs, and the capability to pinpoint m6A sites at single-nucleotide resolution.
In a recent investigation, Liu et al. (2020) harnessed MeRIP-Seq to probe the involvement of m6A modifications in oral squamous cell carcinoma (OSCC). Their findings unveiled pronounced dysregulation in m6A modification profiles within OSCC tissues relative to their normal counterparts, thus underscoring the potential of MeRIP-Seq in deciphering the intricate molecular underpinnings of cancer pathogenesis. Moreover, MeRIP-Seq has been instrumental across diverse studies, elucidating m6A modifications in various biological milieus encompassing embryonic development, cellular differentiation, and the progression of diseases.
Limitations of MeRIP-Seq
While MeRIP-Seq stands as a widely employed method, it is essential to acknowledge certain limitations inherent to its application. Foremost among these is its dependency on immunoprecipitation, a process susceptible to introducing biases and artifacts during the enrichment of RNA. Moreover, the conventional implementation of MeRIP-Seq often necessitates a substantial starting material, thereby potentially constraining its utility in scenarios involving limited RNA quantities. Additionally, it is crucial to recognize that MeRIP-Seq furnishes indirect insights into m6A modification sites and does not afford a comprehensive depiction of the stoichiometry of m6A modifications within individual RNA molecules.
M6A Nanopore Sequencing: Overcoming Challenges
In contrast to MeRIP-Seq, Nanopore sequencing presents a direct and label-free methodology for discerning m6A modifications within RNA molecules. This approach uniquely offers long-read sequencing data, a feature particularly advantageous for probing m6A heterogeneity and temporal dynamics across RNA transcripts.
Pioneering work by Workman et al. (2019) exemplified the efficacy of nanopore sequencing in delineating m6A modifications within human embryonic stem cells (hESCs). Their investigation showcased the heightened sensitivity and specificity of nanopore sequencing in m6A detection, facilitating the direct delineation of m6A modifications at the level of individual RNA molecules. Moreover, nanopore sequencing has found successful application in exploring m6A modifications across diverse biological milieus, spanning plant ontogeny, viral pathogenesis, and oncogenic progression.
| Sequencing Technology | MeRIP-Seq | M6A Nanopore Sequencing |
| Principle | Captures RNA fragments modified with m6A through immunoprecipitation, followed by high-throughput sequencing. | Passes RNA molecules through nanoscale pores and detects ion current changes associated with m6A modification directly. |
| Advantages | - High sensitivity and specificity. - Suitable for low-input RNA samples. - Can identify m6A sites at single nucleotide resolution. | - Direct detection of m6A modification without labeling. - Provides long-read sequencing data. - Enables real-time and single-molecule detection. |
| Limitations | - Relies on immunoprecipitation, which may introduce bias and artifacts. - Requires substantial starting material. - Cannot capture the quantity of m6A modifications on individual RNA molecules. | - Not widely adopted, with challenges in standardization and optimization. - Data analysis is complex and requires specialized bioinformatics support. |
| Applications | - Widely used in the study of RNA m6A modifications. - Applicable to various biological contexts and disease models. | - Holds potential for studying m6A heterogeneity and dynamics. - Suitable for experimental designs requiring long-read sequencing data. |
| Suitability | - Suitable for m6A detection requiring high sensitivity and specificity. - Applicable to experimental designs with sufficient RNA samples. | - Suitable for experimental designs requiring direct detection of m6A modification and long-read sequencing data. - Considerations for sample preparation and data analysis complexity. |
Considerations for Method Selection
When deliberating between MeRIP-Seq and Nanopore sequencing for m6A-seq investigations, researchers must consider multiple variables, encompassing the intricacies of their research inquiries, the attributes of their samples, and the desired sequencing outcomes. MeRIP-Seq may be favored for its standardized protocol, heightened sensitivity, and compatibility with scant RNA inputs. Conversely, nanopore sequencing presents distinctive merits, including direct m6A detection, provision of long-read sequencing data, and single-molecule resolution, rendering it especially pertinent for interrogating m6A heterogeneity and temporal dynamics.
To synthesize, both MeRIP-Seq and Nanopore sequencing constitute invaluable modalities for elucidating m6A modifications within RNA. While MeRIP-Seq retains its status as a cornerstone method endowed with established protocols and superior sensitivity, nanopore sequencing emerges as a direct and label-free alternative offering the promise of real-time, single-molecule detection of m6A modifications. Researchers must judiciously weigh the virtues and constraints of each approach to ascertain the optimal strategy tailored to their experimental requisites.
Conclusion
In summary, m6A-seq stands as a formidable instrument for delving into the involvement of m6A modifications in governing gene expression dynamics and disease etiology. Through harnessing the capabilities of high-throughput sequencing platforms and pioneering methodologies, investigators are poised to unveil the nuanced intricacies underlying m6A modifications and their consequential functional ramifications. In its capacity as a forefront purveyor of genomic solutions, CD Genomics persists in its dedication to propelling the domain of epigenetics forward via state-of-the-art technologies and exhaustive analyses.
References:
- Capitanchik, C., Toolan-Kerr, P., Luscombe, N. M., & Ule, J. (2020). How Do You Identify m6A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets. Frontiers in Genetics, 11, 398.
- Fan, C., Ma, Y., Chen, S., Zhou, Q., Jiang, H., & Zhang, J. (2021). Comprehensive Analysis of the Transcriptome-Wide m6A Methylation Modification Difference in Liver Fibrosis Mice by High-Throughput m6A Sequencing. Frontiers in Cell and Developmental Biology, 9, 767051.
- Zhu, H., Yin, X., Holley, C. L., & Meyer, K. D. (2022). Improved Methods for Deamination-Based m6A Detection. Frontiers in Cell and Developmental Biology, 10, 888279.
- Liu, H., Begik, O., Lucas, M.C. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10, 4079 (2019).
- Hendra, C., Pratanwanich, P.N., Wan, Y.K. et al. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods 19, 1590–1598 (2022).


 Sample Submission Guidelines
Sample Submission Guidelines
