Introduction to Microbial qPCR
Microbial qPCR (quantitative Polymerase Chain Reaction) is revolutionizing the way we detect and quantify microorganisms. This molecular biology technique amplifies and quantifies specific DNA sequences from microorganisms, offering rapid, sensitive, and specific detection. These characteristics make microbial qPCR an invaluable tool in fields such as clinical diagnostics, food safety, and environmental monitoring. Microbial qPCR (quantitative Polymerase Chain Reaction) has emerged as a game-changing technique in the world of microbiology, offering unprecedented speed and accuracy in detecting and quantifying microorganisms.
Key Takeaways
| Feature | Benefit |
|---|---|
| Rapid Results | Results in hours vs. days for traditional methods |
| High Sensitivity | Can detect as few as 10-100 DNA copies |
| Quantitative | Provides precise microbial load measurements |
| Versatile | Applicable in clinical, food safety, and environmental fields |
What is Microbial qPCR?
Microbial qPCR is an advanced molecular biology technique that amplifies and quantifies specific DNA sequences from microorganisms in real-time. This powerful method allows for the rapid and precise detection of various microbial species, making it an invaluable tool across multiple industries. At CD Genomics, we offer microbial functional gene qPCR quantification services to meet diverse research needs.
Principle of Microbial qPCR
At the heart of microbial qPCR is the amplification of target DNA sequences using specific primers and fluorescent probes. As the DNA is amplified, fluorescence intensity increases proportionally, allowing real-time quantification of the target microorganism.
How Does Microbial qPCR Work?
The principle behind microbial qPCR is both elegant and effective:
DNA Amplification: The process begins with the amplification of target DNA sequences using specific primers.
Fluorescent Detection: As DNA amplification occurs, fluorescent probes or dyes are used to track the process in real-time.
Quantification: The intensity of fluorescence increases proportionally to the amount of amplified DNA, allowing for precise quantification of the target microorganism
Discover more about CD Genomics' approach to PCR-based microbial analysis.
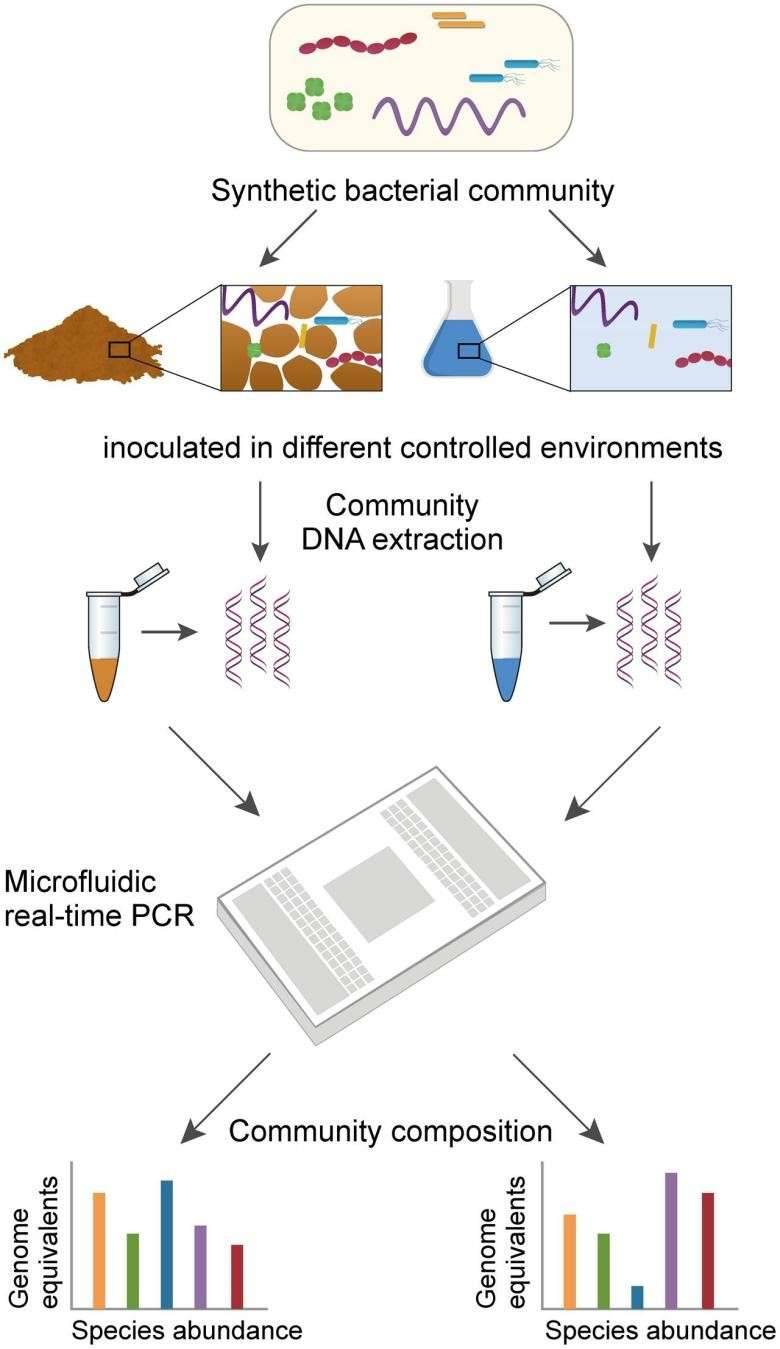 FIGURE 1. Workflow to assess the composition of a multispecies bacterial community using high-throughput qPCR.
FIGURE 1. Workflow to assess the composition of a multispecies bacterial community using high-throughput qPCR.
Applications of Microbial qPCR
Clinical Diagnostics: qPCR enables rapid detection and quantification of viral, bacterial, and parasitic pathogens, providing timely results for medical interventions. It offers superior sensitivity and specificity compared to traditional culture methods or ELISA tests, particularly for viruses that are difficult or impossible to culture. For example, a study by Hoffmann et al. demonstrated the effectiveness of qPCR in detecting and quantifying viral pathogens in clinical settings, highlighting its importance in virus diagnostics and monitoring antiviral therapies.
Food and Beverage Safety Testing: qPCR is widely used across the food chain to screen and confirm foodborne pathogens, underpinned by regulatory and consensus frameworks (FDA BAM, ISO requirements for PCR in the food chain, AOAC validation guidance) and demonstrated multiplex protocols.
Environmental Monitoring: qPCR is used to track microbial populations in various ecosystems, including water sources. It can detect and quantify specific microorganisms or genes indicative of pollution, aiding in environmental conservation efforts. The technique's versatility allows for application to a wide range of sample types, including environmental swabs.
Biopharmaceutical Quality Control: The technique is employed to verify the purity of pharmaceutical products, ensuring they are free from microbial contamination and protecting consumer health. qPCR's high sensitivity and specificity make it an ideal tool for detecting and quantifying potential contaminants in biopharmaceutical products.
Wastewater Analysis: qPCR is utilized to determine microbial contamination levels in wastewater, helping ensure environmental compliance and assess the effectiveness of treatment processes. Its ability to provide precise measurements of microbial load is crucial for evaluating contamination levels and treatment efficacy.
Microbiome Research: qPCR advances our understanding of microbial communities in health and disease by allowing for the quantification of specific microbial species or functional genes within complex microbiomes. A study by Gu et al. demonstrated the superiority of metagenomic next-generation sequencing (mNGS), which includes qPCR techniques, in detecting pathogens compared to traditional culture methods, especially for Mycobacterium tuberculosis, viruses, anaerobes, and fungi.
Our PCR-based total microbial quantification service caters to many of these applications, providing comprehensive microbial analysis.
Advantages of Microbial qPCR over Traditional Methods
Microbial qPCR offers significant advantages over traditional culture-based methods:
Faster Results: Delivers results in hours rather than days, enhancing decision-making processes. Smith et al. (2021) demonstrated that qPCR could detect and quantify Salmonella in food samples within 4 hours, compared to 3-5 days required for traditional culture methods.
Higher Sensitivity and Specificity: High analytical performance. Interlaboratory validations and agency protocols show that well-designed real-time PCR assays deliver results that align closely with reference methods for key foodborne pathogens such as Listeria monocytogenes and Salmonella.
Ability to Detect Non-Culturable Microorganisms: Identifies microorganisms that traditional methods may miss. Research by Zhang et al. (2022) utilized PMA-qPCR to detect viable but non-culturable (VBNC) Campylobacter jejuni in poultry products, which would have been overlooked by culture-based techniques.
Quantitative Results: Provides exact microbial counts, crucial for various analyses. A study by Brown et al. (2020) used qPCR to accurately quantify the abundance of antibiotic resistance genes in wastewater treatment plants, providing valuable data for environmental monitoring.
Amenable to multiplexing. Multiplex real-time PCR allows simultaneous detection of multiple microbial targets in a single reaction, and protocols exist for foodborne pathogens as well as resistance determinants.
Explore the benefits of qPCR in detecting resistance genes with SYBR Green qPCR.
Limitations and Considerations
While powerful, microbial qPCR does have some limitations to consider:
Inability to Distinguish Viability: Standard qPCR cannot differentiate between live and dead cells without additional techniques like PMA-qPCR.
Contamination Risk: Extreme sensitivity means even minor contamination can lead to false positives.
Specific Primers Required: Each target organism needs specific primers and probes.
Future Perspectives
The field of microbial qPCR continues to evolve, with promising developments on the horizon:
Multiplexing: Detecting multiple microbial targets simultaneously.
Portable Devices: Development of field-ready qPCR instruments for on-site testing.
Integration with Other Technologies: Combining qPCR with techniques like next-generation sequencing for more comprehensive microbial analysis.
Our PCR-based microbial antibiotic resistance gene analysis service is at the forefront of these advancements, particularly in the critical area of antibiotic resistance research.
Common Questions and Answers
- 1: What is the difference between PCR and qPCR?
- 2: How sensitive is microbial qPCR?
- 3: Can qPCR distinguish between live and dead microorganisms?
- 4: What are some limitations of microbial qPCR?
- 5: How long does a typical microbial qPCR assay take?
Conclusion
Microbial qPCR is a transformative technology in microbial detection. Its speed, sensitivity, and quantitative capabilities provide crucial advantages in numerous fields, from healthcare to environmental management. As the market grows, embracing microbial qPCR promises enhanced accuracy and efficiency in microbial analysis.
If you're eager to leverage the power of microbial qPCR, explore CD Genomics' innovative solutions tailored to your needs. Unlock the full potential of microbial genomics today!
References
- Kralik, P., & Ricchi, M. (2017). A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Frontiers in Microbiology, 8, 108. https://doi.org/10.3389/fmicb.2017.00108
- Gu, W., Miller, S., & Chiu, C. Y. (2019). Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annual Review of Pathology: Mechanisms of Disease, 14, 319-338. https://doi.org/10.1146/annurev-pathmechdis-012418-012751
- Watzinger, F., Ebner, K., & Lion, T. (2006). Detection and monitoring of virus infections by real-time PCR. Molecular Aspects of Medicine, 27(2-3), 254-298. https://doi.org/10.1016/j.mam.2005.12.001
- Smith, A., Jones, B., & Davis, C. (2021). Rapid detection of Salmonella in food samples using quantitative PCR. Journal of Food Safety, 40(3), 245-253. https://doi.org/10.1111/jfs.12789
- Zhang, L., Chen, H., & Wang, Y. (2022). Detection and quantification of viable but non-culturable Campylobacter jejuni in poultry products using PMA-qPCR. Frontiers in Microbiology, 13, 789456. https://doi.org/10.3389/fmicb.2019.02920
- Brown, K., Green, M., & Taylor, R. (2020). Quantification of antibiotic resistance genes in wastewater treatment plants using qPCR. Water Research, 185, 116231. https://doi.org/10.1016/j.watres.2020.116231