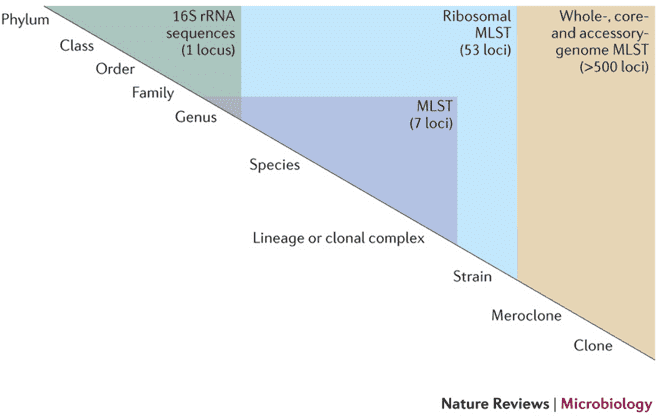
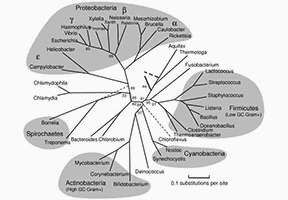
Microbial Diversity and Phylogenetic Tools
A constant obstacle in biology is measuring the complexity of life. In microbiology, the process is confounded by the fact that the census topics are not recognizable to the naked eye or morphologically quickly distinguished, and are projected to number more than 1030 individual bacteria globally. In order to perform a survey of prokaryotes, the characteristics of microorganisms require the use of indirect assessment, which included cultured or 16S rRNA gene sequence assessment. Initial studies range from 10^7to 10^9 for the number of bacterial species in the world. Even though it is well recognized that the number of prokaryotic species in the world is enormous and that the attempts to test them have been insufficient, there has been no comprehensive examination to evaluate how well the bacterial world has been recorded.
To many microbiologists, quantifying microbial phylogenetic diversity is inherently fascinating, but it also plays a key role in the functional analysis of bacterial diversity. Information on the extent of phylogenetic diversity may show how many functional groups have not been compensated for to date. For instance, 16S rRNA diversity studies of terrestrial and marine ecosystems indicated that more than 25 percent of 16S rRNA sequences were gene sequences relating to the Acidobacterium phylum and the SAR11 clade of the -Proteobacteria, respectively. These findings have resulted in enhanced methods of cultivation being developed.
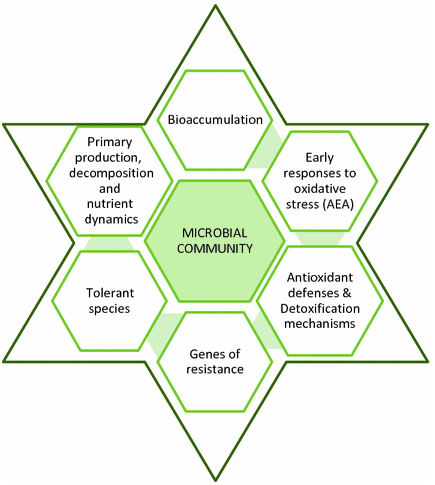
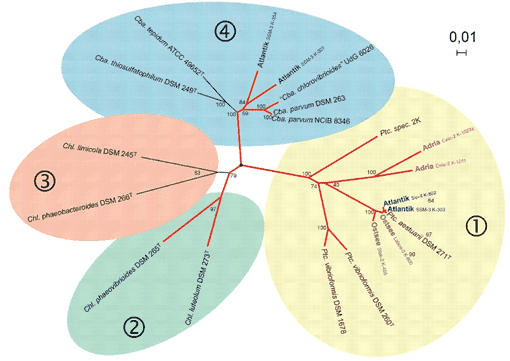 Figure 1. Functional genes in studies to unravel the biodiversity and role of functional microbial groups in the environment. (Imhoff, 2016)
Figure 1. Functional genes in studies to unravel the biodiversity and role of functional microbial groups in the environment. (Imhoff, 2016)
The 16S rRNA gene was suggested as a phylogenetic method to define the evolutionary interactions between organisms and its usage in the classification of unculturable microorganisms in the environment, with GenBank depositing over 78,000 16S rRNA gene sequences. These include sequences separated from cultured bacteria and those amplified without previous cultivation directly from environmental specimens. Sequences acquired by direct environmental amplification offer the only accessible data for 99 percent of prokaryotes in most natural populations. Previous studies have displayed that there are at least 50 bacterial phytates, half of which are made up completely of uncultivated bacteria. There are less than 10% cultured members in an extra three phyla and more than 90% cultured representatives in six phyla.
Microbial Interactions and Networks
Being part of natural ecosystems, microbial communities are innately complicated. Conventional microbiological instruments, such as pure cultures and genetic analysis, appear to offer a reductionist perspective, researching each species in isolation. Nevertheless, for learning about interrelationships and emerging characteristics of populations, a reductionist strategy is not well-suitable.To help infer the connections, holistic methods that straightforwardly investigate natural habitats can produce supplementary info. Trends of co-occurrence also show how specific life forms happen together in a system and differ with environmental variables. These trends demonstrate essential specifics of a precise community structure and can be portrayed as diagrams or networks of mathematical interaction. When environmental circumstances are incorporated in the patterns of co-occurrence, the outcomes imply which situations the organisms' co-occurring lineages favor or ignore. The incorporation of measurements of the biogeochemical level may connect groups of organisms with particular purposes. These types of results enable us to determine the possible relationships within incredibly complicated and evolving natural communities between life forms and factors of the niches of microorganisms.
Applications of Microbial Community-Structure Studies
A number of timely concerns can be addressed using the microbial community framework:
1. Discovering simple microbial transmission trends, like the degree of endemism and ubiquity, and knowing how to assess them in view of the massive 'rare biosphere' and challenges classifying distribution.
2. Looking for general trends that could pertain to all lives and supporting the idea of 'universal' rules that regulate biology in general, thus enhancing our theories and resulting in predictions.
3. To help describe possibly interacting lifeforms and interaction systems in highly complicated systems, using co-occurrence patterns.
4. Using organisms' co-occurrence and environmental variable probabilities to help define microbial species.
5. Assessing repeating trends seasonally or annually, which also results in predictions.
6. Assessing the functions of viruses
7. Connecting community framework or specific genes to particular purposes.
References
- Imhoff JF. New dimensions in microbial ecology—functional genes in studies to unravel the biodiversity and role of functional microbial groups in the environment. Microorganisms. 2016 Jun;4(2).
- Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009 May;459(7244).