Introduction to Microbial Biomarker Detection
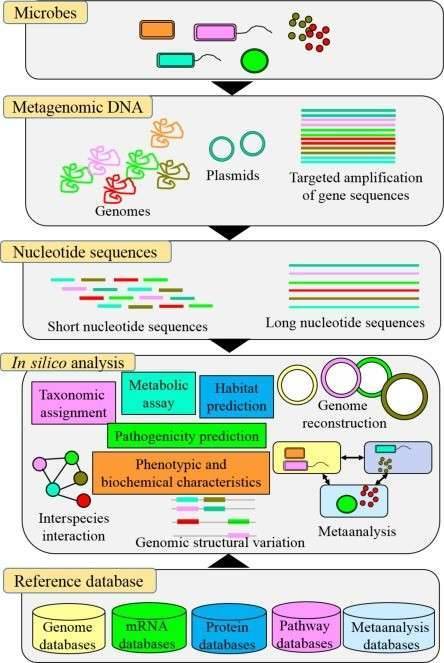
In addition to molding host immunity and hindering infectious disease, commensal microbes linked with humans or animals contribute to the host's health. Because of the prevalence of antimicrobial resistance, promoting a healthy microbiota in humans and animals has become a worldwide concern Biomarker discovery has proven to be one of the most effective and widely applicable methods for converting molecular and genomic data into clinical practice. Even within the same genus, however, the bacterial genome is highly dynamic. As a result, identifying clinically useful biomarkers in mixed microbial communities is extremely difficult. For efficient microbial biomarker discovery, high-resolution techniques such as next-generation sequencing (NGS) and long-read sequencing are required. Deep sequencing, literature-based data mining, and comparative genomics are all examples of genomic methods that can be utilized to systematically explore potential biomarker genes.
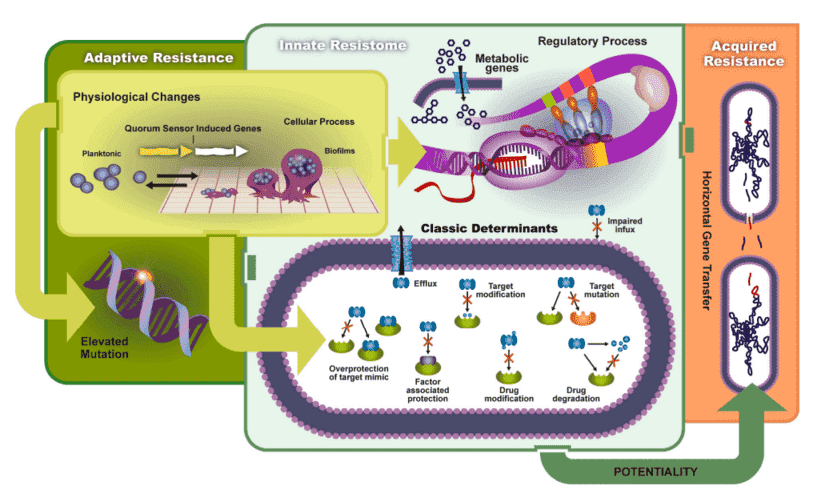
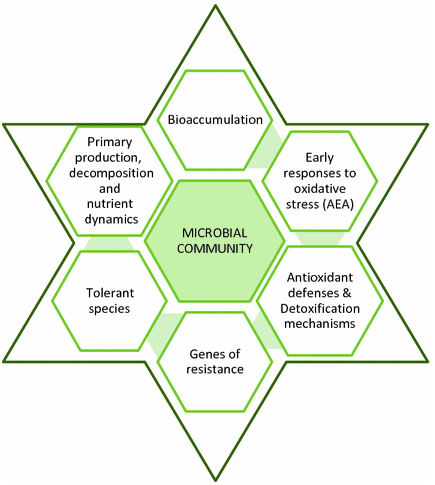 Figure 1. What can microbial biomarkers tell you. (Guasch, 2017)
Figure 1. What can microbial biomarkers tell you. (Guasch, 2017)
Microbial diversity analysis using (full-length) 16S/18S/ITS sequencing
In bacterial taxonomic studies, 16S rDNA has been the most widely utilized molecular clock. It has 9 variable areas and 10 constant areas in its sequence. The degree of variability is closely related to the phylogeny of bacteria, and the variable areas differ with bacteria. Bacterial diversity in the environmental specimen can be studied by identifying sequence variants and abundance of 16S rDNA. As a result, 16S rDNA sequencing is crucial in microbial classification and identification, as well as micro-ecological research.
The 18S rDNA, also known as the ITS (Internal Transcribed Spacer), is widely used to classify and identify fungi. In phylogenetic studies, 18S rDNA is better for phylogenetic level or higher classification; ITS is a relatively conserved area and can be employed to learn species and sub-classic categorical steps.
Discovery of microbial biomarkers using microbial diversity analysis and metatranscriptomics
The study of the function and activity of the entire set of transcripts from environmental specimens is known as metatranscriptomics. On a holistic level, it uncovers the transcripts of metagenomes and the transcriptional regulation rule in specific circumstances and/or time periods. Metatranscriptomics has the advantage of avoiding the isolation and culture of specific microbial species, which efficiently widens the accessible space of resources.
Further categorization of bacteria and fungi using microbial whole-genome sequencing and microbial epigenomics
With high accuracy, whole-genome resequencing can detect DNA biomarkers like single nucleotide polymorphisms (SNPs), insertions and deletions (indels), structure variations (SVs), copy number variations (CNVs), and other genetic changes in the sequenced species. It also offers a once-in-a-lifetime opportunity to characterize polymorphic variants in a population, revealing the underlying processes of species origin, development, growth, and evolution in great detail. Whole-genome re-sequencing is a critical component of genome-wide association studies (GWAS), in which common genetic variants in different individuals are evaluated to see if they are linked to a specific phenotype. Food safety, agriculture, and pharmacy, particularly personalized medicine, can all benefit from GWAS.
DNA methylation, on the other hand, is crucial for the regulation of gene expression, virulence, and pathogen-host interactions. DNA methylation also plays a role in cellular differentiation and development, which is important for understanding gene expression and silencing in cancer and other diseases. Microbial epigenomics sheds light on important tumorigenic pathways, allowing us to identify molecular markers in a variety of biological processes and decipher the mechanisms behind complex diseases. Cell cycle control, DNA repair, developmental biology, and cancer research, and the finding of possible biomarkers and drug targets are all areas where microbial epigenomics analysis can be useful.
References
- Deng X, Li Z, Li G, et al. Comparison of microbiota in patients treated by surgery or chemotherapy by 16S rRNA sequencing reveals potential biomarkers for colorectal cancer therapy. Frontiers in microbiology. 2018 Jul 17;9.
- Shah MS, DeSantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018 May 1;67(5).
- Guasch H, Bonet B, Bonnineau C, Barral L. Microbial biomarkers. InMicrobial Ecotoxicology 2017 (pp. 251-281). Springer, Cham.
- Zautner AE, Masanta WO, Weig M, et al. Mass Spectrometry-based PhyloProteomics (MSPP): A novel microbial typing Method. Scientific reports. 2015 Aug 25;5(1).