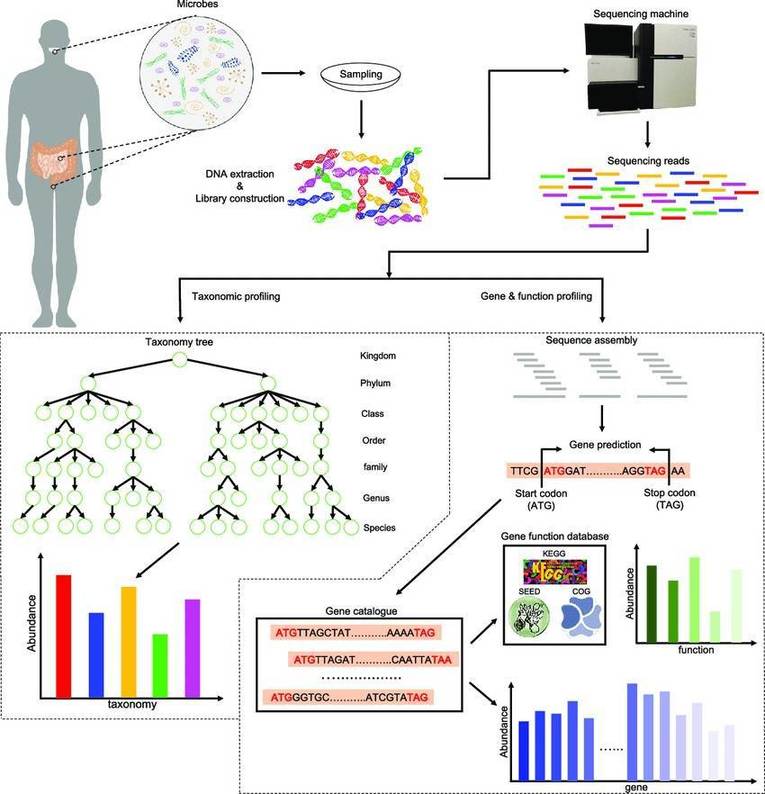
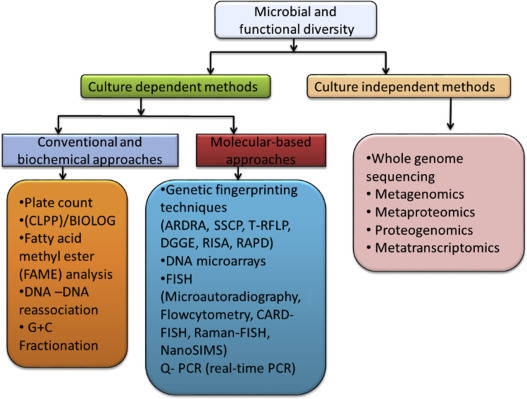
Metagenomics has emerged as a rapidly evolving field of research in recent years, with an increasing volume of studies and a concomitant surge in the data generated. In general, metagenomic studies predominantly encompass two major categories: species composition analysis and functional composition analysis.
This article aims to provide a detailed examination of the functional annotations typically performed in metagenomic research, along with the specific significance of each functional annotation.
KEGG Functional Annotation
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a comprehensive database designed for the systemic analysis of gene products, specifically focusing on their roles within cellular metabolic pathways. This database plays a crucial role in facilitating the study of genes and their expression data within a broader network context. KEGG integrates a wide range of data across several domains, including genomic sequences, chemical molecules, and biochemical systems. Notable categories within KEGG include metabolic pathways (PATHWAY), drugs (DRUG), diseases (DISEASE), gene sequences (GENES), and complete genomes (GENOME).
The KEGG database serves as an invaluable resource for elucidating the functional attributes of genes and their interactions within cellular and organismal systems. Through the KEGG pathways, researchers are able to gain insights into the metabolic networks in which genes are involved, thus enabling a more holistic understanding of biological processes at the molecular level. Additionally, the database provides a means to link gene functions with associated diseases and potential therapeutic interventions, making it indispensable for advancing both basic and applied research in genomics and biotechnology.
 Functional annotation for metagenomic and metabolomic analysis. (Chengpei Zhu et al,.2024)
Functional annotation for metagenomic and metabolomic analysis. (Chengpei Zhu et al,.2024)
CAZy Functional Annotation: Carbohydrate-Active Enzymes Database
The Carbohydrate-Active Enzymes Database (CAZy) is a specialized resource focused on enzymes that catalyze the degradation, modification, and biosynthesis of carbohydrates. This database classifies enzymes into several families based on their functional capabilities, providing valuable insights into the mechanisms of carbohydrate metabolism.
CAZy encompasses five major categories of carbohydrate-active enzymes:
1. Glycoside Hydrolases (GHs): Enzymes that catalyze the hydrolysis of glycosidic bonds between carbohydrate units, facilitating the breakdown of complex carbohydrates.
2. Glycosyltransferases (GTs): Enzymes responsible for transferring sugar moieties from activated donor molecules to specific acceptor substrates, playing a crucial role in the biosynthesis of oligosaccharides and polysaccharides.
3. Polysaccharide Lyases (PLs): Enzymes that cleave glycosidic linkages in polysaccharides through mechanisms that involve the formation of a covalent intermediate, often participating in the degradation of complex polysaccharides like pectins and alginates.
4. Carbohydrate Esterases (CEs): Enzymes that hydrolyze ester bonds in carbohydrate derivatives, thus modifying the structure and properties of polysaccharides, such as those found in plant cell walls.
5. Auxiliary Activities (AAs): A group of enzymes that assist in the degradation of carbohydrates by providing oxidative reactions, such as lignin and polysaccharide oxidation, which often complement the activities of other carbohydrate-active enzymes.
Additionally, CAZy includes a classification for Carbohydrate-Binding Modules (CBMs), which are non-catalytic protein domains that facilitate the binding of enzymes to specific carbohydrate structures, thereby enhancing the efficiency of enzymatic reactions.
The CAZy database is essential for understanding the diversity and functional roles of carbohydrate-active enzymes across various organisms, from microorganisms to plants and animals. By categorizing enzymes based on their mechanistic functions, CAZy offers a comprehensive platform for researchers investigating carbohydrate metabolism, enzyme engineering, and the biotechnological applications of these enzymes.
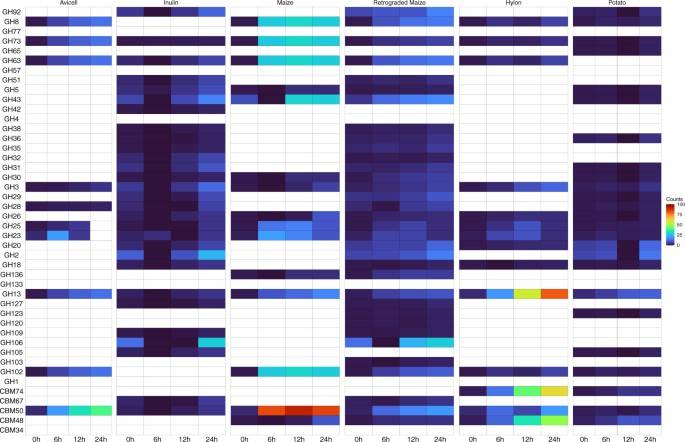 Abundance of extracellular secretory CAZymes. (Anuradha Ravi et al,. 2022)
Abundance of extracellular secretory CAZymes. (Anuradha Ravi et al,. 2022)
PHI Functional Annotation: Pathogen-Host Interactions Database
The Pathogen-Host Interactions (PHI) Database is a publicly accessible and freely available resource that compiles experimentally validated or literature-reported genes associated with pathogenicity, virulence, and effector proteins of pathogens, including fungi, oomycetes, bacteria, and other microbes that infect plants, animals, fungi, and insects. Additionally, the database includes information on antifungal compounds and their corresponding target genes.
COG Functional Annotation: Clusters of Orthologous Groups
COG (Clusters of Orthologous Groups) annotation is a method used for the functional annotation of differential genes. The COG database, developed and maintained by NCBI, classifies gene products based on sequence homology and represents one of the earliest databases for identifying orthologous genes. It is constructed through extensive sequence comparisons across multiple organisms. The COG database is divided into two main categories: one for prokaryotes, referred to as the COG database, and one for eukaryotes, referred to as the KOG database. By comparing protein sequences, a given sequence can be annotated to a specific COG cluster. Each COG cluster consists of orthologous sequences, which allows for the inference of the sequence's function. The COG database is functionally classified into twenty-six categories.
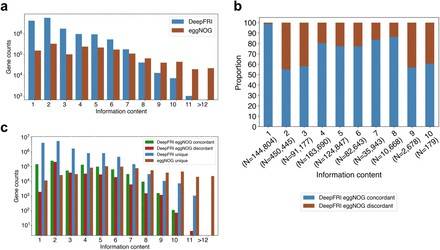 Concordance between DeepFRI and eggNOG annotations.(Mary Maranga et al,.2022)
Concordance between DeepFRI and eggNOG annotations.(Mary Maranga et al,.2022)
The eggNOG (evolutionary genealogy of genes
Non-supervised Orthologous Groups) database is an extension of the NCBI's COG database. It incorporates a broader range of species and a more extensive collection of protein sequence data. The database performs homologous gene clustering and provides descriptions and functional classifications for each homologous gene group. It includes protein sequences from the complete genomes of 5,090 organisms (encompassing eukaryotes, representative bacteria, and archaea) as well as 2,502 viral species. These organisms are categorized into 379 taxonomic groups, with each group assigned a classification number according to NCBI taxonomy. The eggNOG database contains 4.4 million orthologous groups (OGs).
TCDB (Transporter Classification Database) Annotation of Membrane Transport Proteins
The Transporter Classification Database (TCDB) is a comprehensive resource for the classification of membrane transport proteins, a major class of membrane proteins that facilitate the exchange of chemical substances and signals across biological membranes. The lipid bilayer, which surrounds cells and organelles, forms a hydrophobic barrier that isolates the internal environment from the external milieu. While some small molecules can passively diffuse through the membrane, the majority of hydrophilic compounds—such as sugars, amino acids, ions, and drugs—require specialized transporter proteins to traverse this hydrophobic barrier.
Consequently, transporter proteins play a pivotal role in numerous cellular functions, including nutrient uptake, metabolic waste export, and signal transduction. The TCDB (http://www.tcdb.org/) serves as a database for classifying membrane transport proteins, encompassing various families such as the ABC transporter family and the AAA-ATPase transporter family.
Pfam Protein Family Database Annotation
The Pfam database, an abbreviation for "Protein family," provides classification information on protein domain families and is widely used for querying domain annotations and performing multiple sequence alignments. Each protein domain family in Pfam is represented by multiple sequence alignments and Hidden Markov Models (HMMs). Pfam-A, a subset of the database, is constructed using the most recent UniProtKB protein sequence data and is manually curated and validated, ensuring high reliability of its domain annotations. The pfam_scan tool, available on the Pfam website, allows users to analyze protein sequences for the presence of specific domains.
Swiss-Prot Annotation
Swiss-Prot is a curated, non-redundant protein sequence database that is part of the UniProt repository. As of the end of 2018, it included 558,898 protein sequences. Swiss-Prot provides extensive annotations for each protein sequence, including details on protein function, post-translational modifications, structural domains and binding sites, secondary and tertiary structures, and disease associations related to protein defects. These annotations offer valuable insights into the biological roles and functional mechanisms of the proteins.
CARD (Comprehensive Antibiotic Resistance Database) Annotation
The Comprehensive Antibiotic Resistance Database (CARD) is structured around the Antibiotic Resistance Ontology (ARO), which serves as the primary classification framework for the database. ARO consists of terms that link antibiotic modules to their targets, resistance mechanisms, gene variants, and other related information. Additionally, CARD offers a specialized software tool called the Resistance Gene Identifier (RGI), designed to predict resistance genes within genomic data based on the functional annotations of ARO. This tool aids in the identification and analysis of antibiotic resistance genes across various genomes.
 The Comprehensive Antibiotic Resistance Database provides data, models, and algorithms relating to the molecular basis of antimicrobial resistance. (From Wikipedia)
The Comprehensive Antibiotic Resistance Database provides data, models, and algorithms relating to the molecular basis of antimicrobial resistance. (From Wikipedia)
BacMet (Antibacterial Biocide and Metal Resistance Genes Database) Annotation
The BacMet database (Antibacterial Biocide and Metal Resistance Genes Database) contains a comprehensive collection of 753 genes associated with resistance to metal ions and biocides. This resource allows for the identification of metal ion and biocide resistance genes within target strains, as well as the identification of genes that confer dual resistance to both types of stressors, making it highly valuable for resistance research. The database categorizes genes based on their functional roles, grouping metal resistance genes and biocide resistance genes according to their resistance mechanisms and evolutionary relationships. This classification is particularly significant for data mining in environmental microbial communities, such as those found in various soil microbiomes and anaerobic activated sludge, providing essential guidance for studying microbial resistance in different ecological contexts.
VFDB (Virulence Factors of Pathogenic Bacteria) Annotation
Virulence factors (VFs) are molecules produced by bacteria, viruses, fungi, and other pathogens that possess invasive and toxic properties, enabling them to infect host organisms. These factors facilitate pathogen entry into host tissues and cells by suppressing or evading the host immune response, thereby allowing the pathogen to acquire nutrients and proliferate. Virulence factors may be encoded within mobile genetic elements, such as plasmids, genomic islands, and bacteriophages, and can be horizontally transferred between bacteria, transforming harmless strains into virulent pathogens. Therefore, when identifying virulence factors, particular attention is often given to genomic islands, secreted proteins, and other related elements.
MetaCyc (Metabolic Pathways From All Domains of Life) Database Annotation
MetaCyc is a non-redundant database of experimentally elucidated metabolic pathways, encompassing both primary and secondary metabolism. It includes detailed information on associated metabolites, reactions, enzymes, and genes, and is widely used in plant metabolomics. Currently, MetaCyc contains 3,153 pathways, 19,020 reactions, and 19,372 metabolites, with continuous updates. As an online metabolic encyclopedia, MetaCyc can be utilized to predict metabolic pathways in sequenced genomes and supports metabolic engineering through its integration with enzyme databases.
antiSMASH (Antibiotics and Secondary Metabolite Analysis Shell) Database Annotation
antiSMASH is a tool designed for the identification of secondary metabolite gene clusters. Typically, genes encoding biosynthetic enzymes involved in secondary metabolism are clustered on chromosomes. Utilizing profile hidden Markov models (HMMs) for specified types, antiSMASH accurately identifies all known secondary metabolite gene clusters. In the antiSMASH database, these gene clusters are classified into 24 categories. The most common secondary metabolite gene clusters include type I, II, and III polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS). For example, tetracyclines, macrolides, ansamycins, and polyethers are synthesized via the PKS pathway, while beta-lactams, polypeptides, and glycopeptides are synthesized through the NRPS pathway.
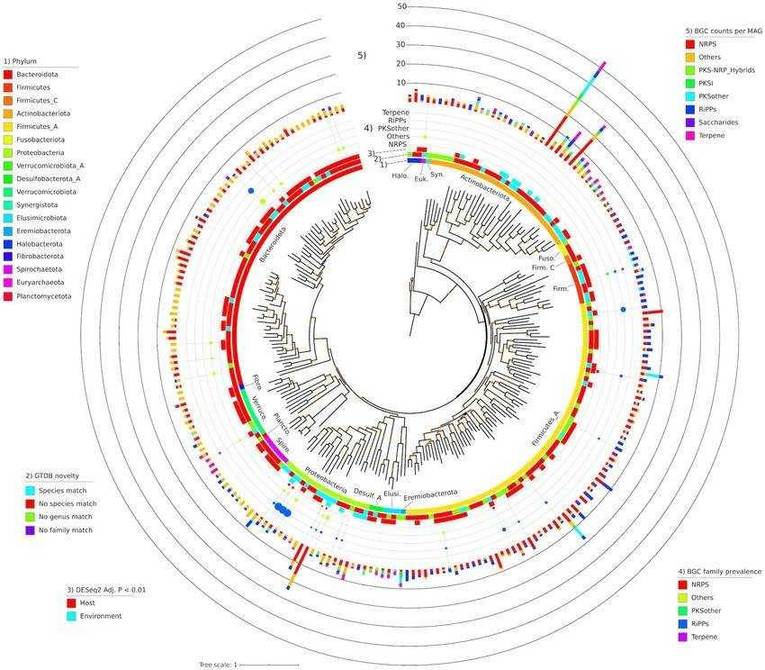 Phylogeny of all SGBs (n 233) with 3 BGCs identified by AntiSMASH. (Nicholas Youngblut et al,.2020)
Phylogeny of all SGBs (n 233) with 3 BGCs identified by AntiSMASH. (Nicholas Youngblut et al,.2020)
Services you may interested in
Resource
Summarizes and compares the functional annotation of various databases
| Database | Focus | Key Features | Categories/Functions | Applications |
|---|---|---|---|---|
| KEGG (Kyoto Encyclopedia of Genes and Genomes) | Cellular metabolic pathways | Systemic analysis of gene products and metabolic pathways | Metabolic pathways, drug interactions, diseases, genes, complete genomes | Understanding biological processes, metabolic networks, and therapeutic interventions |
| CAZy (Carbohydrate-Active Enzymes Database) | Carbohydrate-active enzymes | Classification of enzymes involved in carbohydrate degradation, modification, and biosynthesis | Glycoside Hydrolases (GHs), Glycosyltransferases (GTs), Polysaccharide Lyases (PLs), Carbohydrate Esterases (CEs), Auxiliary Activities (AAs) | Carbohydrate metabolism, enzyme engineering, biotechnology applications |
| PHI (Pathogen-Host Interactions Database) | Pathogen-host interactions | Genes associated with pathogenicity, virulence, and effector proteins | Pathogenicity, virulence, antifungal compounds, host-pathogen interactions | Pathogen identification, disease research, microbial ecology |
| COG (Clusters of Orthologous Groups) | Orthologous gene groups | Functional annotation based on sequence homology | 26 functional categories including metabolism, genetic information, transport | Gene functional inference, comparative genomics, evolutionary studies |
| eggNOG (Evolutionary Genealogy of Genes) | Orthologous gene groups | Expanded COG database with more species and protein sequences | Orthologous groups (OGs), functional classification | Homologous gene clustering, gene function prediction |
| TCDB (Transporter Classification Database) | Membrane transport proteins | Classification of membrane transport proteins | ABC transporters, AAA-ATPases, ion channels | Studying membrane transport mechanisms, cellular nutrient uptake |
| Pfam (Protein Family Database) | Protein domain families | Protein family classification, multiple sequence alignments, Hidden Markov Models | Protein domains, functional annotation | Sequence alignment, protein structure prediction |
| Swiss-Prot | Protein sequence annotation | Curated, non-redundant protein sequence database | Protein function, post-translational modifications, disease associations | Protein function analysis, disease-related research |
| CARD (Comprehensive Antibiotic Resistance Database) | Antibiotic resistance genes | Classification of antibiotic resistance mechanisms and gene variants | Resistance mechanisms, gene variants | Antibiotic resistance prediction, genomic data analysis |
| MGE (Mobile Genetic Element Database) | Mobile genetic elements (MGEs) | Elements involved in horizontal gene transfer and genome evolution | Insertion sequences (IS), transposons, integrons, plasmids, phages | Horizontal gene transfer studies, gene mobility, environmental adaptation |
| BacMet (Antibacterial Biocide and Metal Resistance Genes Database) | Biocide and metal resistance genes | Resistance genes associated with biocides and metals | Resistance to biocides, resistance to metals | Microbial resistance studies, environmental microbiome analysis |
| VFDB (Virulence Factors of Pathogenic Bacteria Database) | Virulence factors of pathogens | Genes responsible for pathogenicity and virulence in bacteria | Pathogenicity, virulence, secretion systems | Identification of virulent strains, virulence factor research |
| MetaCyc (Metabolic Pathways from All Domains of Life) | Metabolic pathways | Non-redundant metabolic pathways for primary and secondary metabolism | Metabolic pathways, reactions, enzymes, metabolites | Metabolomics, metabolic pathway prediction, enzyme engineering |
| antiSMASH (Antibiotics and Secondary Metabolite Analysis Shell) | Secondary metabolite gene clusters | Identification of biosynthetic gene clusters for secondary metabolites | Polyketide Synthases (PKS), Non-Ribosomal Peptide Synthases (NRPS) | Secondary metabolite research, antibiotic biosynthesis, microbial genome analysis |