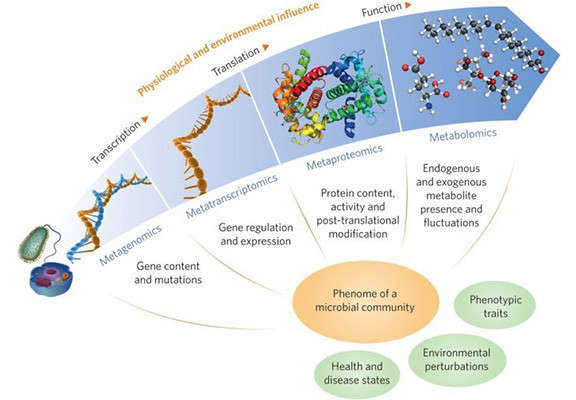
Gene expression analysis helps scientists understand how cells grow, function, and react to their environment. It plays a big role in studying diseases, finding new treatments, and learning how the body works. In this review, we explain three main technologies: microarrays, RNA sequencing (RNA-Seq), and quantitative real-time PCR (qPCR). We describe how each method works, their strengths and limitations, and the situations where they are best used. We also highlight key applications in research and medicine and discuss future directions in this fast-moving field.
1. Overview of Gene Expression Analysis
Gene expression tells us which genes are active in a cell at a given time. By studying these patterns, scientists learn how cells develop, respond to stress, and fight diseases. It also helps us find mistakes in gene activity that may cause illnesses like cancer or genetic disorders.
In the early days, measuring gene expression was slow and focused on one gene at a time. Methods like Northern blotting could only study a few genes and took a lot of effort. Today, with technologies like microarrays and RNA-Seq, researchers can measure thousands of genes at once. These new tools allow for a deeper and more detailed understanding of biology.
At the same time, computer tools for analyzing gene expression data have become much stronger. Bioinformatics helps manage, process, and interpret the large volumes of data produced by modern experiments. As a result, gene expression analysis has become a key part of biology, medicine, and biotechnology.
2. Main Technologies for Gene Expression Analysis
Several powerful technologies are now widely used to measure gene expression. Each method offers different advantages depending on the research goals.
Microarray Analysis
Microarrays use a solid surface with thousands of tiny DNA probes. Researchers add fluorescently labeled RNA or cDNA from their samples. If the sample matches a probe, it binds and gives off a fluorescent signal. Special scanners read these signals and measure how much of each RNA is present.
Microarrays allow researchers to study many genes at once. They are great for comparing different samples, like healthy vs. cancerous tissues. Scientists have used microarrays to find gene signatures that predict how patients will respond to treatment.
However, microarrays have some limits. They can only detect genes that have matching probes on the array. Also, they are not very good at spotting new, unexpected RNA forms. Despite these limits, microarrays are still popular because they are affordable, fast, and reliable for many standard tasks.
RNA Sequencing
RNA-Seq uses next-generation sequencing (NGS) to read the sequences of RNA molecules. First, RNA is broken into smaller pieces. Then it is copied into complementary DNA (cDNA) and special adapters are added. These cDNA fragments are sequenced, producing millions of short reads. Computers match these reads to a reference genome or assemble them into full transcripts.
RNA-Seq provides a complete picture of gene expression. It can detect rare transcripts, new RNA types, alternative splicing, and mutations. Unlike microarrays, RNA-Seq does not need prior knowledge of the genes being studied.
New versions of RNA-Seq, such as single-cell RNA-Seq and spatial transcriptomics, allow scientists to study gene expression in individual cells or in their original tissue context. These methods are helping researchers understand the complexity of tissues and uncover rare cell types that were previously missed.
Quantitative Real-Time PCR
qPCR is a highly accurate method for measuring specific gene expression. Scientists design primers and fluorescent probes that match their genes of interest. As the PCR reaction amplifies the target DNA, the fluorescent signal increases. By measuring this signal in real time, researchers can calculate how much starting RNA was present.
qPCR is fast, sensitive, and works well for both high and low levels of gene expression. It is often used to confirm findings from larger studies, such as microarray or RNA-Seq experiments. A newer version called digital PCR splits the sample into thousands of small reactions, allowing even more precise measurements, especially useful for rare transcripts.
3. Gene Expression Analysis with qPCR
Despite the rise of high-throughput technologies, qPCR remains the gold standard for targeted gene expression studies.
Workflow
RNA Extraction and Reverse Transcription: First, high-quality RNA must be carefully extracted. Then, reverse transcription enzymes turn RNA into cDNA.
Primer and Probe Design: Researchers design short DNA sequences (primers) and fluorescent probes that specifically bind to their target genes.
PCR Amplification and Detection: During the PCR reaction, fluorescence grows as the target DNA is amplified. The machine measures fluorescence at each cycle.
Data Analysis
Ct Value: The cycle threshold (Ct) is the number of cycles needed for fluorescence to cross a set threshold. A lower Ct means more starting RNA.
Relative Quantification: Comparing the Ct of target genes to stable reference genes using the ΔΔCt method allows comparison between samples.
Absolute Quantification: Standard curves based on known amounts of RNA help determine exact molecule numbers.
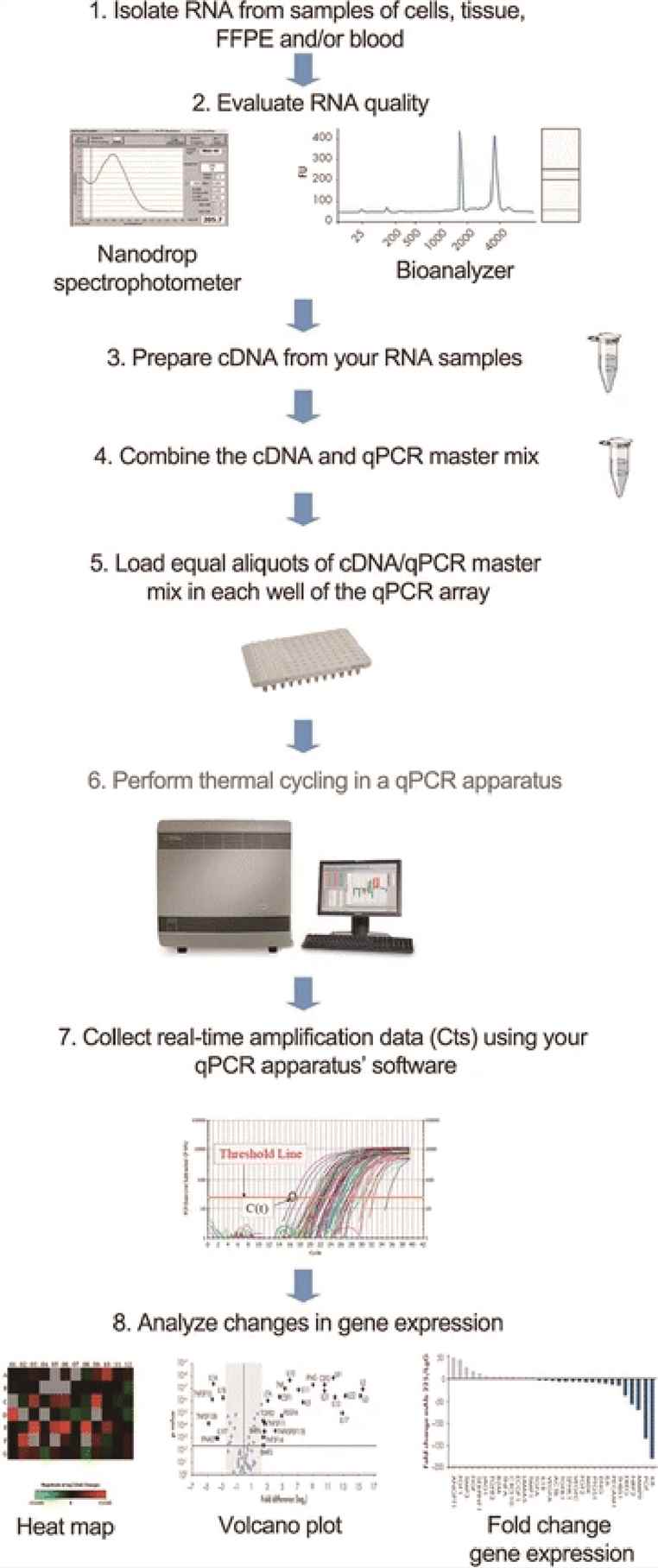 Figure 1 . Workflow of qPCR analysis.(Alvarez ML. et al, 2014)
Figure 1 . Workflow of qPCR analysis.(Alvarez ML. et al, 2014)
Advantages and Limitations
qPCR is very sensitive and specific. It requires small amounts of RNA and gives results quickly. However, it can only test a few genes at a time, making it less suitable for discovery studies.
4. Gene Expression Analysis with Microarrays
Microarrays marked the start of genome-wide expression studies and are still widely used today.
Workflow
Sample Labeling: RNA is converted into labeled cDNA using fluorescent dyes.
Hybridization: Labeled cDNA is applied to the array and binds to matching probes.
Washing and Scanning: After washing off unbound material, a laser scanner detects the fluorescent signals.
Data Analysis
Normalization: Methods like quantile normalization remove technical biases between arrays.
Statistical Testing: Linear models and Bayesian approaches find genes with significant expression changes.
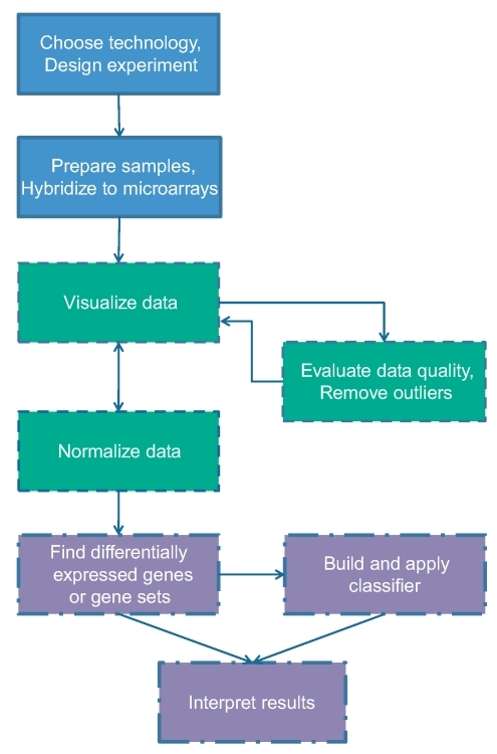 Figure 2 . Workflow of Microarrays analysis.(Slonim DK. et al, 2009)
Figure 2 . Workflow of Microarrays analysis.(Slonim DK. et al, 2009)
Advantages and Limitations
Microarrays are cost-effective and good for studying large sample sets. They provide reproducible results when studying known genes. However, they have a limited dynamic range and cannot detect unknown RNAs.
5. Gene Expression Analysis with RNA-Seq
RNA-Seq has become the most powerful tool for studying gene expression across the entire transcriptome.
Workflow
Library Preparation: RNA is fragmented, converted to cDNA, and adapters are added.
Sequencing: High-throughput sequencers read millions of short sequences.
Mapping and Assembly: Reads are aligned to a reference genome or assembled without one.
Data Analysis
Quantification: Expression levels are usually reported as TPM (Transcripts Per Million) or FPKM (Fragments Per Kilobase of transcript per Million mapped reads).
Differential Expression: Statistical tools identify genes that change between conditions.
Discovery: RNA-Seq can uncover novel transcripts, fusion genes, and alternative splicing events.
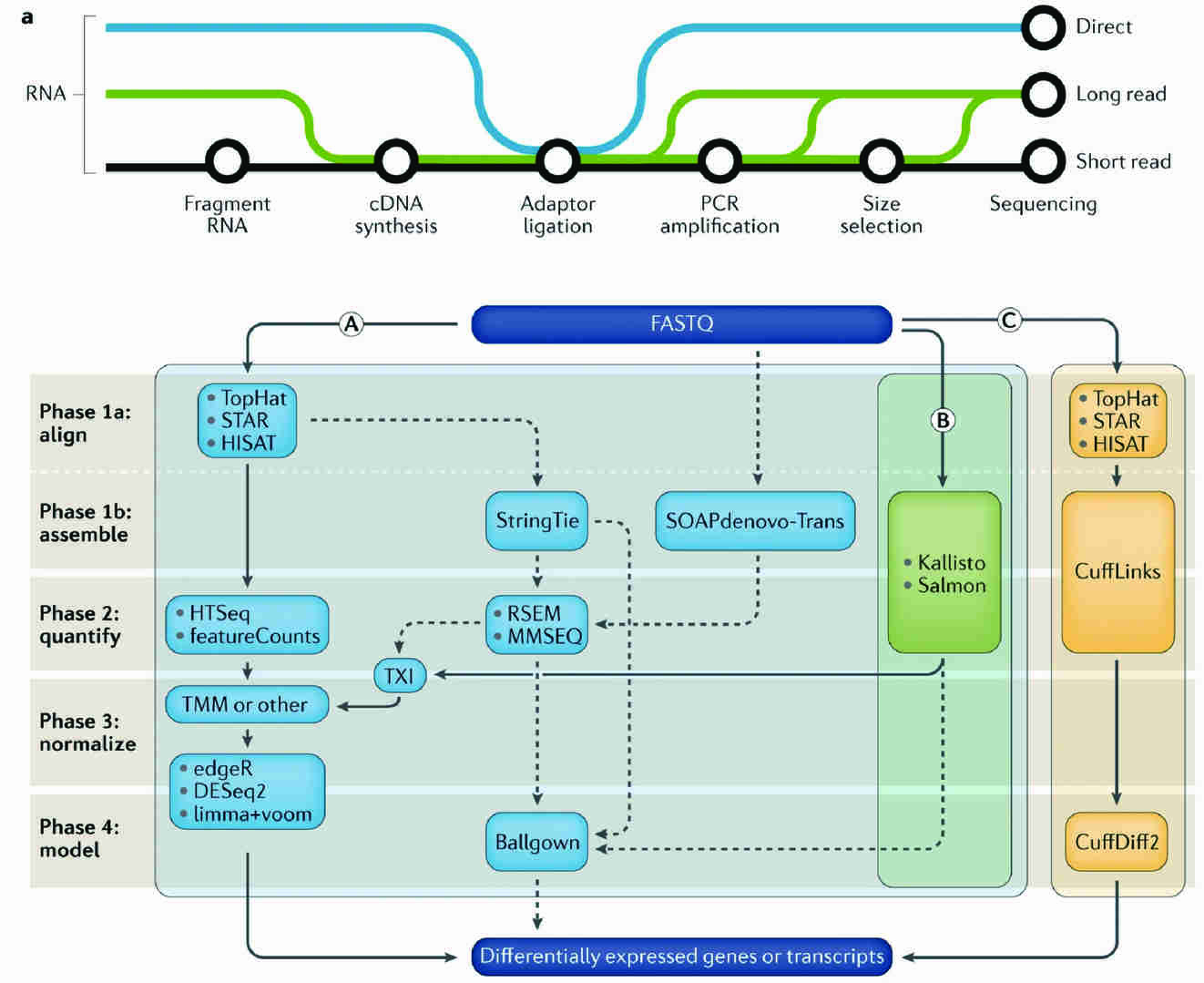 Figure 3 . Workflow of RNA-seq analysis.(Stark R. et al, 2019)
Figure 3 . Workflow of RNA-seq analysis.(Stark R. et al, 2019)
Advantages and Limitations
RNA-Seq offers unmatched detail and flexibility. It captures a wide dynamic range and detects new RNA forms. However, it is more expensive and complex than microarrays and requires strong bioinformatics support.
Recent innovations include long-read sequencing technologies, like PacBio and Oxford Nanopore, which can sequence full-length RNA molecules. This helps resolve complex splicing patterns and structural variations missed by short-read methods.
6. Comparing Gene Expression Analysis Technologies
Similarities
All three methods measure RNA levels and help uncover biological insights. Each fits different research needs depending on cost, complexity, and study goals.
Differences
Microarrays: Best for profiling known genes across large sample sets.
RNA-Seq: Best for exploring unknown regions, rare transcripts, and complex samples.
qPCR: Best for highly accurate measurements of specific genes.
Choosing the Right Method
- For discovery projects, RNA-Seq is the top choice.
- For validating findings or focused studies, qPCR is preferred.
- For large epidemiological studies where cost matters, microarrays are practical.
Sometimes, researchers use a combination-broad profiling with RNA-Seq or microarrays, followed by targeted validation with qPCR.
7. Gene Expression Analysis Applications in Research and Medicine
Gene expression analysis is used widely across biology and healthcare:
Cancer: Expression profiles help classify tumors, predict outcomes, and guide treatments. Tests like Oncotype DX in breast cancer are based on gene expression.
Immunology: Studying immune cell gene expression reveals how the body fights infections or develops autoimmune diseases.
Neuroscience: Researchers uncover gene patterns involved in brain development, learning, memory, and disorders like autism or Alzheimer's disease.
Pharmacogenomics: Gene expression helps predict drug responses and identify patients at risk for side effects.
Infectious Disease: RNA signatures from patients' blood can distinguish between bacterial and viral infections quickly.
New trends like cell-free RNA testing are making it possible to monitor health and disease with just a blood sample, without needing a biopsy.
8. Future Directions
Gene expression analysis is moving toward even higher resolution and broader applications:
Single-Cell Technologies: Allow study of individual cells, revealing hidden diversity in tissues.
Spatial Transcriptomics: Preserve spatial context of gene expression in tissues, helping understand organ organization and tumor microenvironments.
Integration with Other Omics: Combining expression data with genomics, proteomics, and metabolomics provides a fuller view of biology.
Artificial Intelligence: Machine learning tools are improving pattern recognition and predictions based on expression data.
As costs drop and technologies improve, gene expression analysis will become even more central to biology, precision medicine, and biotechnology innovation.
9. Conclusion
Gene expression analysis has revolutionized biological research. Technologies like microarrays, RNA-Seq, and qPCR each have unique strengths. Choosing the right method depends on research goals, available resources, and the need for discovery or validation.
With ongoing advances in sequencing, single-cell studies, and computational methods, gene expression analysis will continue to drive major discoveries in health and disease. Understanding and applying these powerful tools is essential for the next generation of scientific breakthroughs.
References
- Alvarez ML et al., SYBR® Green and TaqMan® quantitative PCR arrays: expression profile of genes relevant to a pathway or a disease state. Methods Mol Biol. 2014;1182:321-59. https://doi.org/10.1007/978-1-4939-1062-5_27
- Slonim DK et al., Getting started in gene expression microarray analysis. PLoS Comput Biol. 2009 Oct;5(10):e1000543. https://doi.org/10.1371/journal.pcbi.1000543
- Stark R et al., RNA sequencing: the teenage years. Nat Rev Genet. 2019 Nov;20(11):631-656. https://doi.org/10.1038/s41576-019-0150-2