Service Specifications
Introduction to Our Microbial DNA Metabarcoding Service
CD Genomics offers comprehensive DNA metabarcoding services aimed at investigating the microbial diversity within environmental samples. Utilizing our Illumina next-generation sequencing platform, we provide reliable and in-depth microbial DNA barcoding and sequencing services. These capabilities allow for the precise identification of microorganisms across various taxonomic levels and facilitate the comparative analysis of microbial diversity among multiple samples.
Our advanced sequencing platforms are capable of generating millions of sequences rapidly, thus enabling the efficient study of complex environmental samples. The metabarcoding process involves the use of primers to amplify DNA present in the environment, while high-throughput sequencing is employed to obtain extensive taxonomic information encoded within the genes. We deliver detailed data analysis of numerous sequence fragments derived from complex environmental samples. This analysis primarily includes noise removal, acquisition of operational taxonomic unit (OTU) data, and subsequent analyses based on OTUs.
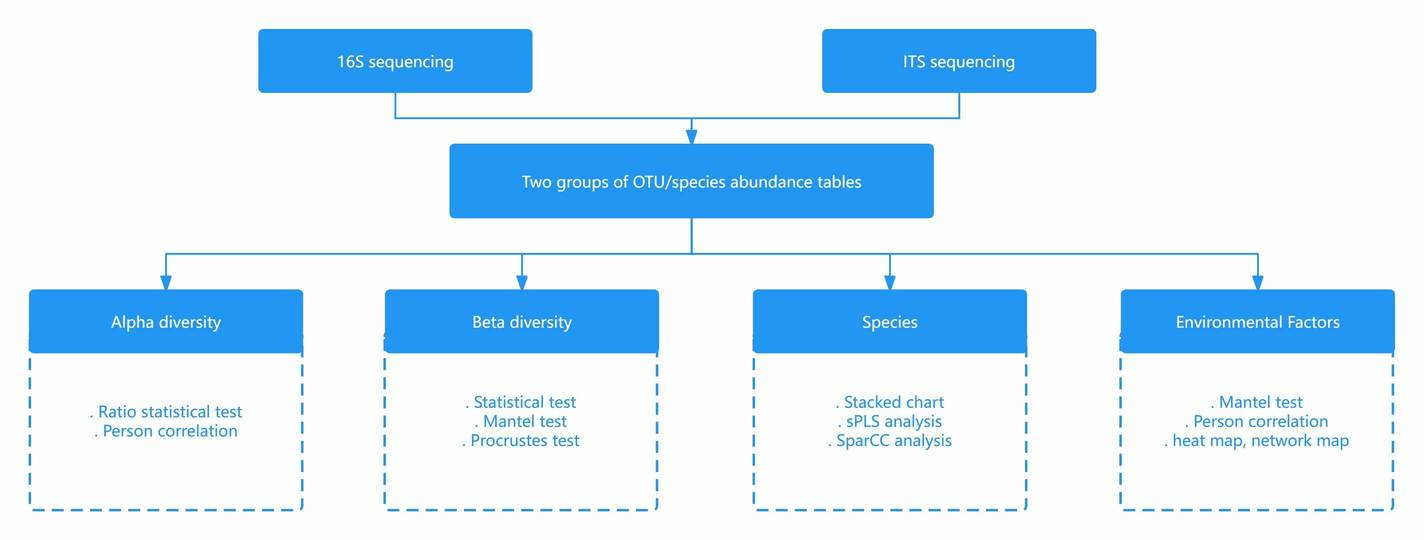
Microbial DNA Metabarcoding Workflow
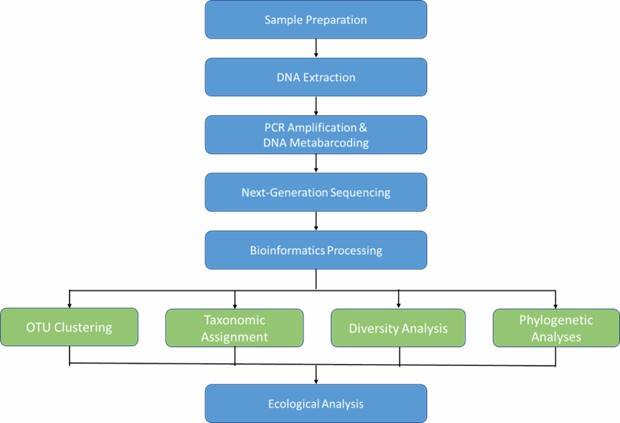
Technical Parameters
DNA Sequencing Strategies:
- For PCR products (target DNA fragments) smaller than 280 bp, use Illumina PE150 sequencing.
- For DNA fragments larger than 280 bp but smaller than 480 bp, use Illumina PE250 sequencing.
- For DNA fragments larger than 480 bp but smaller than 580 bp, use Illumina PE300 sequencing.
- For PCR products larger than 580 bp, use PacBio sequencing.
Note: We design appropriate sequencing strategies based on your plan and objectives, utilizing suitable sequencing platforms. Please feel free to contact us directly.
Bioinformatics Analysis
Our bioinformatics services offer scientists custom analysis to address real issues apart from data quality control, gene alignment, genome assembly, and phylogenetic analyses. Our bioinformatics analysis services provide comprehensive interpretations of environmental DNA (eDNA) metabarcoding. For more details, please refer to the following table.
| Analysis Content |
Details |
| Taxonomic assignment |
Reference-based methods are used for the precise taxonomic assignment of sequencing reads. |
| Metabolic analysis |
Metabolic processes encoded in the genome, from biosynthesis to biodegradation, directly link microbial communities to the environment. |
| Habitat prediction |
Predict possible habitats of microbes. |
| Genome reconstruction |
Reconstruction of microbial genome by metagenomic binning and assembly from various environments. |
| Inter-species interactions |
Use abundant information from large-scale metagenomic datasets, co-occurrences (or anti-occurrences) among microbes, hosts, and/or viruses can be studied. |
| Genomic structural variations |
Observe systematic variations in the gene order (or gene cluster structures) due to gene losses, fusions, duplications, inversions, translocations, and HGTs from an analysis of metagenome data. |
| Meta-analysis |
Meta-analysis of multiple datasets can reveal general patterns or laws that determine how microbes interact with their environments and how their genomes have been shaped. |
Note: The above content includes only a portion of the bioinformatics analysis. For more information or to customize the analysis, please contact us directly.
Sample Requirement
- DNA/cDNA amount ≥ 1μg, Concentration≥10 ng/µL, No obvious degradation, 1.8 < OD260/280 < 2.0, OD260/230 ≥ 1.8,
- Soil: 2–5 g per sample
- Water: More than 1 filter membrane per sample
- Feces: 1–3 g per sample
- Air: More than 2 filter membranes per sample
- Sludge/Sediment: 5–10 g per sample
Note:
If you wish to obtain more accurate and detailed information regarding sample requirements, please feel free to contact us directly.
Deliverables
- Raw sequencing data (FASTQ)
- Clean data
- Trimmed and stitched sequences (FASTA)
- Quality-control dashboard
- Statistic data
- Your designated bioinformatics result report



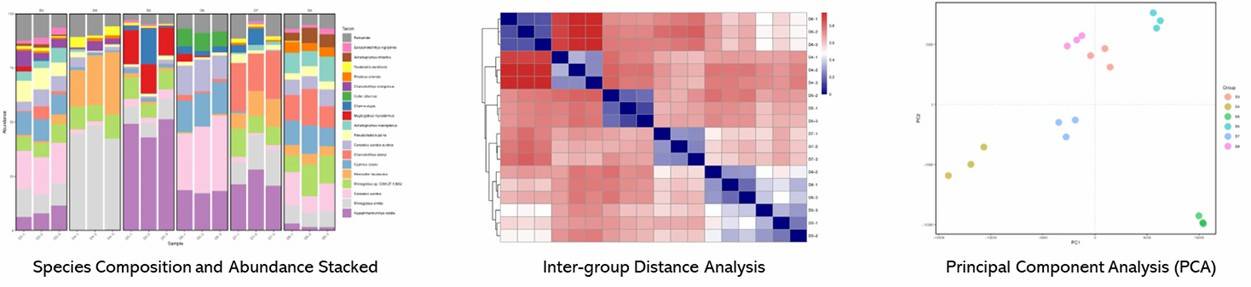

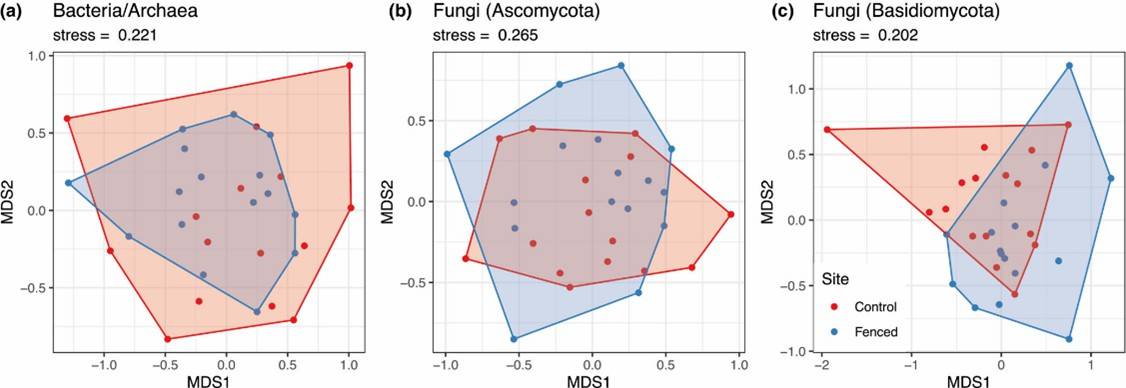 Figure 1. Nonmetric multidimensional scaling plot.
Figure 1. Nonmetric multidimensional scaling plot. Figure 2. Results of functional guild composition analysis for the soil fungal communities.
Figure 2. Results of functional guild composition analysis for the soil fungal communities.