Eubacteria belong to the prokaryotic domain, with prokaryotic cell structure, nuclear wrapped by nuclear membrane, and genetic material DNA exposed in the quasi-nuclear region. Binary fission is the main breeding mode, and some of them can form spores to resist adversity in harsh environment. Metabolic types are diverse, including photoautotrophic, chemoautotrophic, photoheterotrophic and chemoheterotrophic. Respiratory types include aerobic, anaerobic and facultative anaerobic, which adapt to various ecological environments. Scientists have studied them through microscope observation, culture and molecular biology.
This paper provides a comprehensive overview of eubacteria, covering classification, reproduction, metabolism and research methods, and shows its importance and research value in the ecosystem.
The Classification of Eubacteria
Eubacteria can be divided into many groups according to phenotypic characteristics, such as Gram staining and aerobicity. Involving many groups, it provides an important basis for bacterial identification and classification system construction, and promotes the development of related research.
Gram-positive Bacteria
Cladosporium: This kind of bacteria has a thick cell wall. Common Bacillus, such as Bacillus subtilis, widely exists in soil and other environments and can produce spores to resist adverse environment; There are also lactic acid bacteria, such as Lactobacillus acidophilus, which are often used in food fermentation and play a role in yogurt production, which is beneficial to human intestinal health.
Actinomycetes: Most of them grow in hyphae. Streptomyces is a typical representative. streptomyces griseus can produce a variety of antibiotics and make important contributions to the medical field. Frankia can symbiotic nitrogen fixation with non-leguminous plants and enhance the utilization of nitrogen by plants.
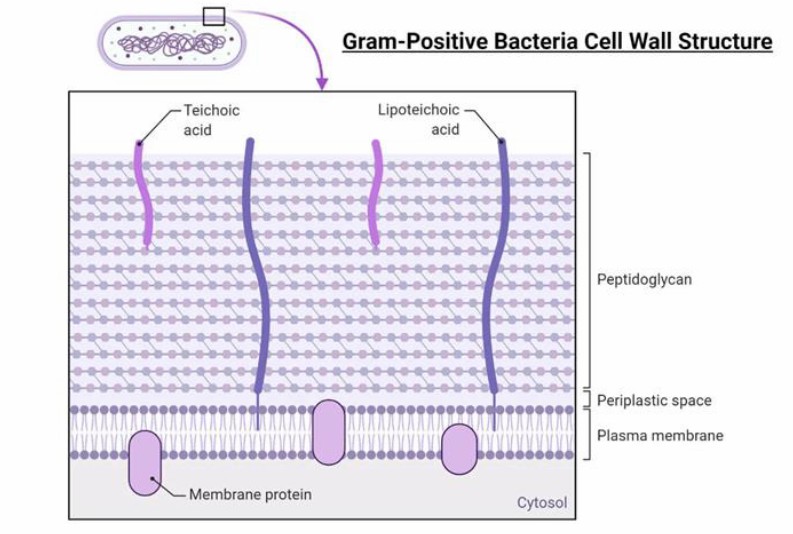
Gram-positive bacteria cell wall structure
Gram-negative Bacteria
Proteus: This is a huge and diverse group. Escherichia coli belongs to Enterobacteriaceae, which is common in human and animal intestines. Some strains are conditional pathogens, while normal strains contribute to intestinal digestion. Vibrio cholerae belongs to Vibrio family, which is the pathogen that causes cholera. It spreads through polluted water and food, seriously threatening human health.
Bacteroides: Bacteroides is relatively common, mostly exists in human and animal intestines, and participates in the process of food decomposition and digestion, which is of great significance for maintaining intestinal microecological balance.
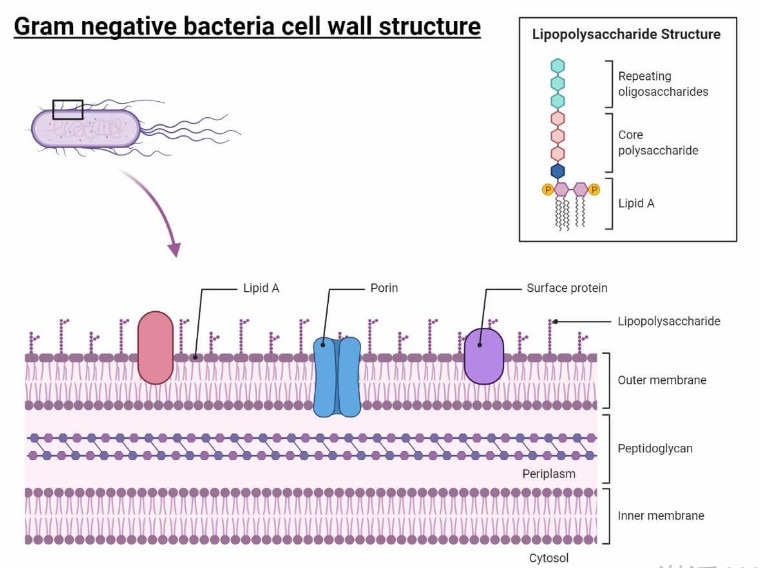
Gram-negative bacteria cell wall structure
Other Categories
Cyanobacteria: It can carry out photosynthesis. For example, Spirulina is rich in nutrients such as protein and is widely developed as a health care product; Nostoc flagelliforme is also a kind of cyanobacteria, which has the function of nitrogen fixation in the ecosystem.
Chlamydia: Chlamydia trachomatis in Chlamydia can cause trachoma and is a common pathogen of eye infection, which is mainly transmitted through contact.
Mycoplasma: Mycoplasma pneumoniae can cause mycoplasma pneumonia, which is easy to spread among people, especially children and adolescents. It has no cell wall and has various forms.
How Does Eubacteria Reproduce
Binary fission: Binary fission is the most important breeding mode of eubacteria. Under suitable environmental conditions, bacterial cells first replicate DNA, which doubles the genetic material in cells. Subsequently, the cell membrane in the middle of the cell was depressed inward, forming a transverse wall, and at the same time, the cell wall began to synthesize and grow to the middle, eventually splitting the cell into two, forming two basically identical daughter cells. This breeding method is simple and efficient, and the breeding speed is extremely fast. For example, under suitable nutrition and temperature conditions, binary fission of E.coli can be completed every 20 minutes or so, and a large number of daughter cells can be produced in a short time, rapidly increasing the population.
Spore formation: When the environmental conditions become bad, such as high temperature, drought, lack of nutrition or invasion of harmful substances, some eubacteria will form spores. Spore is a kind of dormant structure with high stress resistance produced by bacteria under certain conditions. Spores have multi-layer thick walls, extremely low internal water content, almost stop metabolic activities, and have strong resistance to high temperature, ultraviolet rays, drying and chemical disinfectants. For example, spores formed by Bacillus anthracis can survive in soil for years or even decades. Once the environmental conditions are improved, spores will germinate and transform into vegetative cells with metabolic activity and reproductive ability again, which enables bacteria to preserve themselves in extreme environment and wait for the right time to resume growth and reproduction.
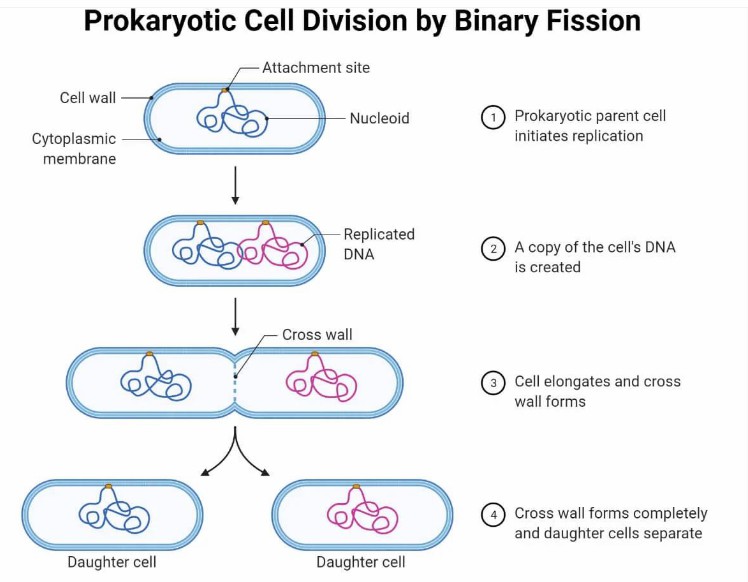
Eubacteria cell devision by binary fission
Different Metabolic Types of Eubacteria
The metabolic modes of eubacteria are extremely diverse, which can be divided into four categories according to different energy and carbon sources. Photoautotrophic uses photoenergy to synthesize organic matter with carbon dioxide; Chemotactic autotrophic oxidized inorganic matter can obtain energy and fix carbon dioxide; Photoenergy heterotrophy uses light energy and takes organic matter as carbon source; Chemical energy heterotrophy relies on oxidation of organic matter to obtain energy and carbon source.
Diversified Nutritional Types
Photoautotrophic: this kind of eubacteria can use light energy as energy source and carbon dioxide as carbon source to synthesize organic substances they need and release oxygen at the same time. Typical representatives, such as cyanobacteria, contain photosynthetic pigments such as chlorophyll a in their cells, which can carry out photosynthesis like plants. Cyanobacteria convert light energy into chemical energy through photosynthesis, which is stored in organic compounds such as sugars, providing energy and material basis for their own growth and reproduction, and playing an important role in carbon cycle and oxygen production in the earth ecosystem.
Chemotactic autotrophic: This kind of eubacteria uses the chemical energy released by oxidizing inorganic substances to fix carbon dioxide to synthesize organic matter. Nitrifying bacteria, for example, can oxidize ammonia to nitrous acid and then to nitric acid, gain energy in the process, and use carbon dioxide to synthesize cell matter. Nitrifying bacteria participate in the nitrogen cycle in nature, which is of great significance for maintaining soil fertility and ecological balance.
Photoenergy heterotrophy: This kind of bacteria use light energy to obtain energy, but they need to take organic matter from the environment as carbon source and hydrogen donor. For example, Rhodosporus can grow by using organic substances (such as organic acids, alcohols, etc.) in the environment under the condition of light, and at the same time use light energy as a supplementary energy source. Photoenergy heterotrophic bacteria occupy a specific niche in the ecosystem and participate in the transformation of organic matter and energy flow.
Chemotactic heterotrophy: Most eubacteria belong to chemotactic heterotrophy, and they rely on oxidizing organic substances (such as sugar, protein, fat, etc.) to obtain energy and carbon sources. The metabolic pathways of chemotactic heterotrophic bacteria are diverse, which can be adjusted according to the types and availability of organic substances in the environment. E. coli, for example, can grow and reproduce by using abundant organic nutrients in the human intestinal environment, and its metabolic activities have an important impact on the microecological balance of the human intestinal tract.
Differences in Respiratory Types
Aerobic respiration: Aerobic eubacteria breathe in an aerobic environment, with oxygen as the final electron acceptor. Through a series of complex enzymatic reactions, organic matter is completely oxidized and decomposed into carbon dioxide and water, and a lot of energy is released. For example, under aerobic conditions, Bacillus subtilis efficiently oxidizes organic substances such as glucose through glycolysis, tricarboxylic acid cycle and oxidative phosphorylation, providing sufficient energy for cell growth and metabolism.
Anaerobic respiration: Anaerobic eubacteria breathe in an anaerobic environment, with inorganic oxides such as nitrate, sulfate and carbonate or some organic compounds as the final electron acceptor. For example, Vibrio desulfurization can use sulfate for anaerobic respiration, reduce sulfate to hydrogen sulfide, and at the same time oxidize organic matter to obtain energy. Although anaerobic respiration produces relatively little energy, it enables bacteria to survive and multiply in a special environment without oxygen.
Facultative anaerobic respiration: Facultative anaerobic eubacteria have a more flexible way of breathing, aerobic respiration when aerobic and anaerobic respiration or fermentation when anaerobic. Taking yeast as an example, under aerobic conditions, yeast carries out aerobic respiration to multiply in large quantities; Under anaerobic conditions, yeast converts sugar into alcohol and carbon dioxide through fermentation, producing a small amount of energy to maintain life activities. This characteristic enables facultative anaerobic bacteria to adapt to many different environmental conditions.
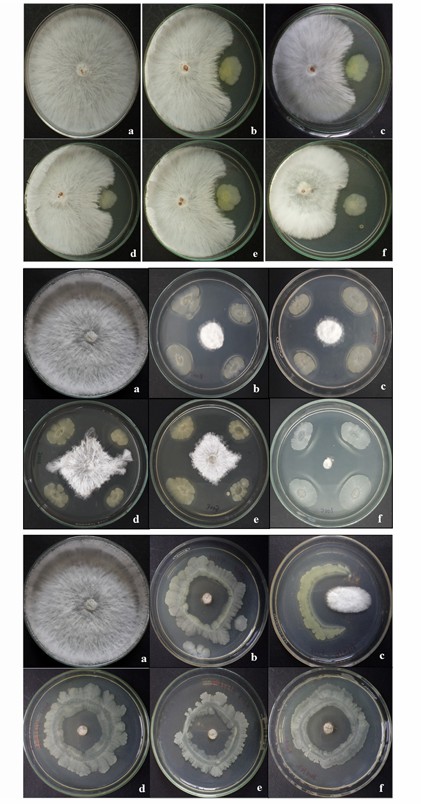
In vitro interaction between bacterial isolates and S. rolfsii on potato dextrose agar media plate (Kumari et al., 2020)
How to Investigate Eubacteria
Eubacteria, as one of the oldest and diverse biological groups on the earth, in-depth study on it is helpful to reveal the mystery of life, promote the development of biotechnology and maintain ecological balance. The following will introduce the research methods of eubacteria from the aspects of sample collection, culture separation, identification technology and research means.
Sample Collection
Environmental selection: Eubacteria are widely distributed in various environments, and different environments need to be selected for different research purposes. If we study soil microbial community, we should select representative soil areas, such as farmland, forest, grassland and so on. When sampling in farmland, crop types, planting years and fertilization are considered, because these factors affect the species and quantity of eubacteria. If you pay attention to the bacteria in water, you can take samples at different depths and locations in rivers, lakes and oceans, and the bacterial communities in surface water and bottom water may be different.
Sampling method: Soil samples are usually collected by layers at a certain depth with a soil drill, and the soil at different depths is mixed into a sample to reflect the overall bacterial situation of the soil. Water sampling can use water sampler, such as plexiglass water sampler, to collect water samples in different water layers. For special environment, such as high-temperature hot springs, special high-temperature resistant sampling equipment is needed to ensure that the survival state of bacteria is not disturbed too much during the collection process.
Storage and transportation: the collected samples should be processed as soon as possible, and if they cannot be processed in time, they should be stored at low temperature. Soil samples can be placed in a refrigerator at 4℃, and water samples can be refrigerated after adding preservatives (such as formaldehyde). During transportation, samples should be sealed to prevent leakage and pollution. For samples in anaerobic environment, special anaerobic sampling bags or containers should be used for transportation.
Culture Separation
Selection of culture medium: Select the appropriate culture medium according to the nutritional requirements of the target bacteria. For most heterotrophic bacteria, beef paste peptone medium can be used; For autotrophic bacteria, such as nitrifying bacteria, inorganic carbon source and nitrogen source should be used as the main culture medium. To isolate bacteria with specific functions, such as cellulase-producing bacteria, cellulose can be added to the culture medium as the only carbon source, so that only bacteria that can decompose cellulose can grow.
Separation methods: Dilution coating plate method and scribing separation method are commonly used. Dilution coating plate method is to coat the sample on the surface of solid culture medium after gradient dilution. After culture, individual bacteria form independent colonies, which can be further purified and cultured. The streaking separation method is to continuously streak the surface of solid culture medium with inoculation ring, so that the bacteria are gradually dispersed and finally form a single colony. For some bacteria that are difficult to culture, the enrichment culture method can be used to culture samples under specific conditions (such as specific temperature and pH value), so that the target bacteria can dominate in the community before separation.
Identification Technology
Morphological identification: Observe the morphology of bacteria by microscope, including cell shape (spherical, rod-shaped, spiral, etc.), size, arrangement (single, paired, chain-shaped, etc.) and whether there are special structures (spores, capsules, flagella, etc.). Gram staining is an important morphological identification method. According to the staining results, bacteria can be divided into Gram-positive bacteria and Gram-negative bacteria. There are differences in cell wall structure and composition between the two types of bacteria, and the staining characteristics are helpful for preliminary classification.
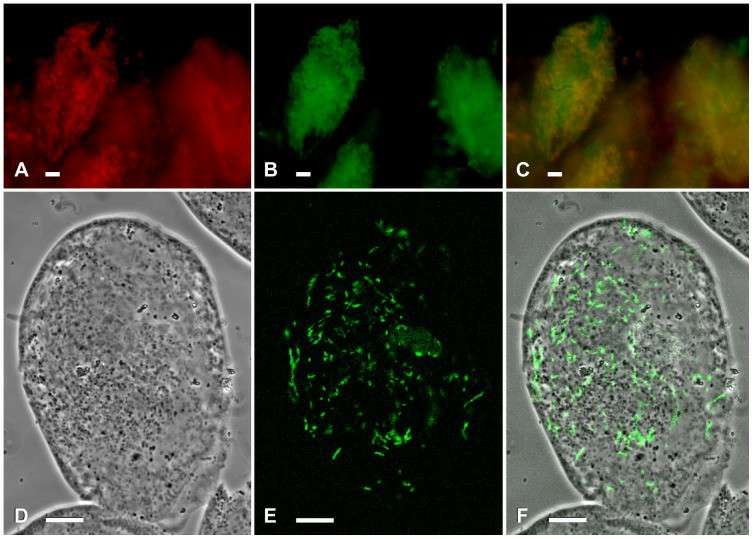
Fluorescence in situ hybridizations of Spirostomum sp. 72 and Euplotes octocarinatus FL(12)-VI (Schrallhammer et al., 2013)
Molecular biological identification: The identification method based on bacterial nucleic acid sequence analysis has high accuracy. The sequencing of 16S rRNA gene is widely used. The 16S rRNA gene exists in all bacteria and has conservative region and variable region. By amplifying and sequencing the variable region sequence and comparing it with the gene database, the taxonomic status of bacteria can be determined. The higher the similarity, the closer the genetic relationship. In addition, genome-wide sequencing can obtain complete genetic information of bacteria and analyze the relationship between bacterial characteristics and evolution more accurately.
Unlike eubacteria, which rely on 16S rRNA sequencing for classification, fungi utilize ITS (Internal Transcribed Spacer) regions. High-throughput ITS2 sequencing can reveal cross-domain microbial networks, such as fungal-bacterial symbiosis in soil ecosystems.
Research Methods
Genomics technology: Whole genome sequencing can analyze bacterial genetic information and understand gene function, metabolic pathway and evolutionary relationship. Transcriptomics studies the gene transcription of bacteria under different conditions and reveals the regulation mechanism of gene expression. Protein omics analyzed the composition and function of bacterial protein, and studied the post-translation modification of protein and the interaction between protein and protein.
Metabonomics technology: Metabonomics detects the variety and content changes of bacterial metabolites. By analyzing the fingerprint of metabolites, we can understand the metabolic state of bacteria and its response to environmental changes. Gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS) are commonly used metabonomics analysis techniques, which can separate and identify complex metabolites.
Ecological research method: The aim is to study the ecological function and relationship of eubacteria in the natural environment. Using fluorescence in situ hybridization (FISH) technology, the distribution and abundance of specific bacteria in environmental samples were detected with fluorescence-labeled nucleic acid probes. An ecological model was constructed to simulate the dynamic changes of bacterial community, and the influence of environmental factors on the structure and function of bacterial community was analyzed.
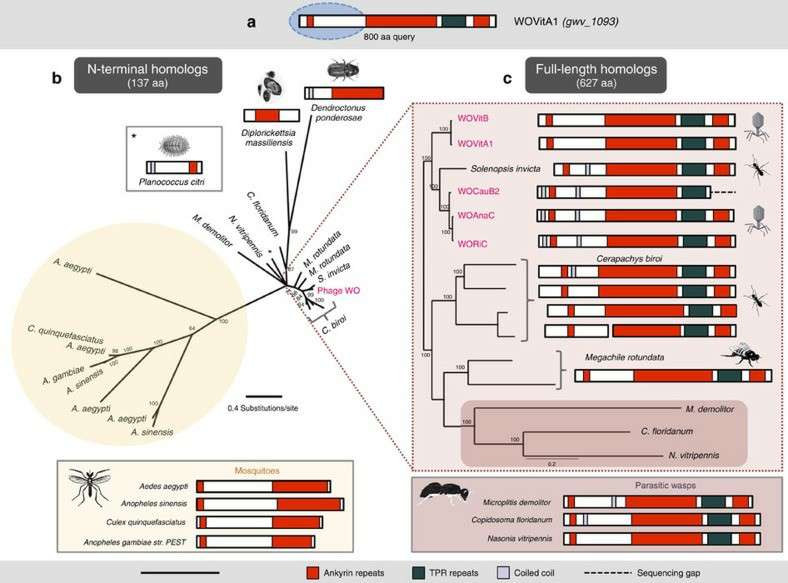
Related TPR and ankyrin proteins support lateral genetic transfer (Bordenstein et al., 2016)
Conclusion
Eubacteria belong to the prokaryotic domain, showing rich diversity and occupying a key position in the ecosystem. In the aspect of reproduction, binary fission is the main way, and the cell replicates the genetic material efficiently and divides it into two parts, thus realizing the rapid growth of population in a short time. When the environment is bad, some true bacteria form spores, which have strong resistance to stress and can help bacteria survive in extreme environment and recover their activity when conditions are suitable.
In terms of metabolic types, eubacteria have photoautotrophic type, which uses photoenergy to fix carbon dioxide. Chemical energy autotrophic, oxidizing inorganic substances to obtain energy; Photoenergy heterotrophic, with the help of light energy and dependent on organic matter. Chemo-heterotrophic, relying on oxidized organic matter to survive, breathing types include aerobic, anaerobic and facultative anaerobic to adapt to different environments. There are various research methods. Microscope is used to observe morphology, culture technology can obtain pure culture, and molecular biology means can help to analyze genes, so as to deeply understand the characteristics, functions and mechanism of eubacteria in the ecosystem.
Learn More:
- Eubacteria: Introduction of Characteristics, Living Environments, Ecosystem and Human Relations
- Unraveling Fungi through Sequencing: Approaches and Applications
References
- Schrallhammer M, Ferrantini F., et al. "’Candidatus Megaira polyxenophila’ gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent rickettsiae." PLoS One. 2013 8(8):e72581 https://doi.org/10.1371/journal.pone.0072581
- Kumari, P., Bishnoi, S.K. Chandra, S. "Assessment of antibiosis potential of Bacillus sp. against the soil-borne fungal pathogen Sclerotium rolfsii Sacc. (Athelia rolfsii (Curzi) Tu & Kimbrough)." Egypt J Biol Pest Control 31, 17 (2021) https://doi.org/10.1186/s41938-020-00350-w
- Bordenstein SR, Bordenstein SR. "Eukaryotic association module in phage WO genomes from Wolbachia." Nat Commun. 2016 7:13155 https://doi.org/10.1038/ncomms13155
- Chiellini C, Pasqualetti C., et al. "Harmful Effect of Rheinheimera sp. EpRS3 (Gammaproteobacteria) Against the Protist Euplotes aediculatus (Ciliophora, Spirotrichea): Insights Into the Ecological Role of Antimicrobial Compounds From Environmental Bacterial Strains." Front Microbiol. 2019 10:510 https://doi.org/10.3389/fmicb.2019.00510
- Quast C, Pruesse E., et al. "The SILVA ribosomal RNA gene database project: improved data processing and web-based tools." Nucleic Acids Res. 2013 (Database issue):D590-6 https://doi.org/10.1093/nar/gks1219
- Ordóñez CD, Redrejo-Rodríguez M. "DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions." Int J Mol Sci. 2023 24(11):9331 https://doi.org/10.3390/ijms24119331


 Sample Submission Guidelines
Sample Submission Guidelines