Summary of CRISPR-Cas9 off-target Detection Methods
Genome editing technology stands at the forefront of current life science research. However, for its successful translation into clinical applications, the accurate detection of off-target effects is indispensable. Assessing and mitigating off-target effects correctly is a pressing concern. The issue of off-target effects in CRISPR-Cas9 gene editing technology has been a longstanding focus of attention. There are two main approaches for examining CRISPR-Cas9 off-target effects: computational simulation methods and sequencing methods [1].
Sequencing Methods of CRISPR-Cas9 off-target Detection
Whole Genome Sequencing (WGS)
Whole Genome Sequencing (WGS) is associated with minimal assessment bias in the context of evaluating Cas9-induced off-target mutations. Currently, sequencing depths of 30-60x are typically employed. However, the cost of WGS is high, and with lower sequencing depths, only a small number of clones may be sequenced, potentially missing the majority of low-frequency off-target events. For in vivo analysis, WGS remains the preferred method, yet techniques that directly capture Double-Strand Breaks (DSBs), such as BLESS, might offer superior capabilities compared to WGS. This is because WGS can also capture naturally occurring variations between cell/animal models, which could complicate analysis and lead to misleading results.
In the analysis of WGS, selecting appropriate control groups is particularly crucial. To minimize false positives and negatives in WGS, the analysis often necessitates the setup of multiple groups, which can be both labor-intensive and expensive.
Chip-Seq
Cas nucleases, a class of proteins, can be subjected to antibody-based pull-down followed by sequencing, a method known as ChIP-Seq. ChIP-Seq is commonly employed in the context of detecting off-target effects associated with dCas9. In contrast to conventional Cas9, which severs the target sequence, leading to its dissociation from the DNA sequence and unstable binding, dCas9, characterized by nuclease activity impairment, retains the ability to bind to the target sequence guided by gRNA. As a result, dCas9 is frequently applied for CRISPR activation (CRISPRa) to augment gene expression rather than for gene knockout.
DISCOVER-seq, also known as MRE11 ChIP-Seq, is a recently developed method for CRISPR-Cas9 off-target sequencing. This approach leverages the recruitment of the endogenous DNA repair factor MRE11 to identify Cas-induced double-strand breaks on the entire genome. During the DNA repair process, MRE11 is precisely recruited to the site of DSB occurrence. Consequently, by pulldown of MRE11-bound sites and high-throughput sequencing, this method allows for the simultaneous examination of both on-target and off-target occurrences. Importantly, DISCOVER-seq is applicable to both in vivo and in vitro samples.
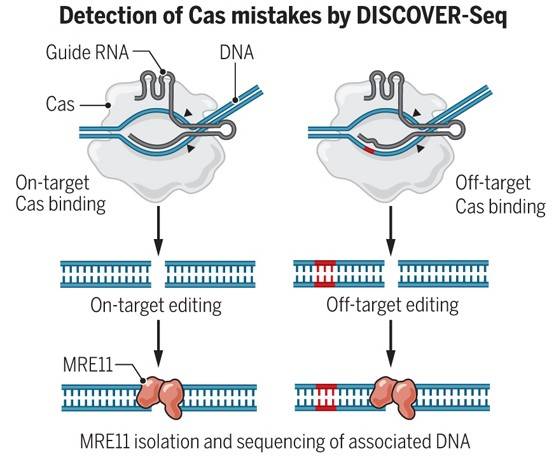 discover-seq
discover-seq
Cas Nuclease Cleavage Site and DSB Site Enrichment Sequencing
The method of sequencing that relies on enriching regions bound by or cleaved by Cas nucleases is commonly referred to as an unbiased detection approach. Following this rationale, enrichment methods have been designed, including:
(1) Antibody Pulldown:
Such as Chip-Seq, where antibodies are used to capture the target regions.
(2)Enrichment of Cas Nuclease Cleavage Sites:
Digenome-Seq, short for in vitro Cas9-digested whole-genome sequencing. After Cas9 cleavage, a 5' phosphate group is left behind, which allows for adapter ligation for PCR amplification and subsequent NGS sequencing. In Digenome-Seq, purified genomic DNA (gDNA) is treated with Cas9 in vitro, followed by adapter ligation, and comprehensive whole-genome sequencing. This method is suitable for in vitro samples.
(3) Enrichment of DNA Double-Strand Break Regions (DSB) - DSB Capture:
In BLESS-Seq, direct biotin affinity enrichment is employed to capture CRISPR-Cas9-guided DSB fragments, which can then be sequenced after adapter ligation. Biotin affinity enrichment specifically captures DSBs that are actively occurring, leaving already repaired DSBs unaccounted for, earning it the label of an "off-target snapshot." This method is commonly applied to cell and tissue samples.
In IDLV-Seq, the integration-deficient lentiviral vector (IDLV) can be used as a PCR anchor to amplify the DSB sites generated through non-homologous end joining (NHEJ). This enables the detection of off-target sites. However, the efficiency of this method relies on the IDLV's ability to integrate into DSBs via NHEJ. Additionally, due to the random integration characteristics of lentiviruses, there is a possibility of false positives.
HTGST is suitable for detecting chromosome translocations induced by CRISPR-Cas9, but it's worth noting that CRISPR-Cas9-induced off-target events primarily result in indels, which are relatively less frequent. Additionally, this method is still subject to limitations imposed by chromatin accessibility.
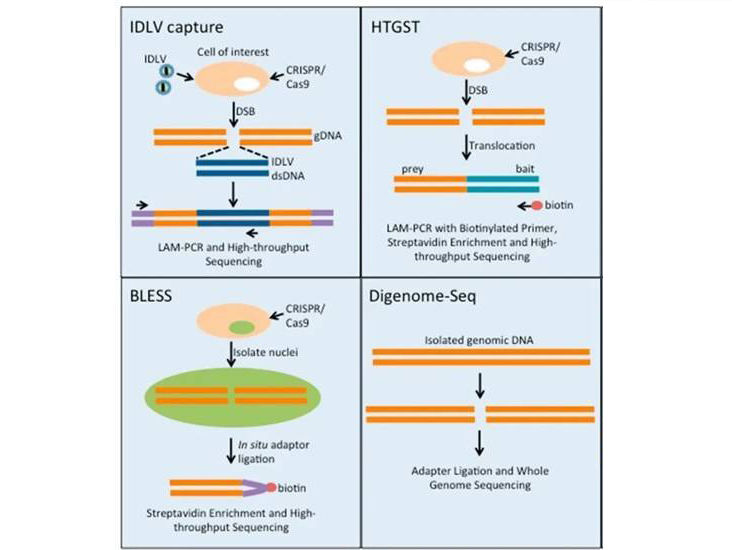 unbiased method
unbiased method
Compared to methods that enrich nucleases' cleavage sites, DSB capture methods are more physiologically relevant. However, they may exhibit lower efficiency since most DSBs are of very short duration. The in vivo tagging of IDLV and the ligation of in situ linkers are both potentially influenced by local chromatin and sequence composition near the cleavage site. Consequently, capture methods may be subject to biases.
Summary of Sequencing Methods
Numerous sequencing methods are available, making it infeasible to provide exhaustive descriptions of each. The table presented below offers the most comprehensive compilation currently accessible. Given the information provided earlier, definitively determining the superior method is not feasible. The selection of the most appropriate sequencing technique should be guided by the specific needs of the experiment and the financial resources at the disposal of the research team.
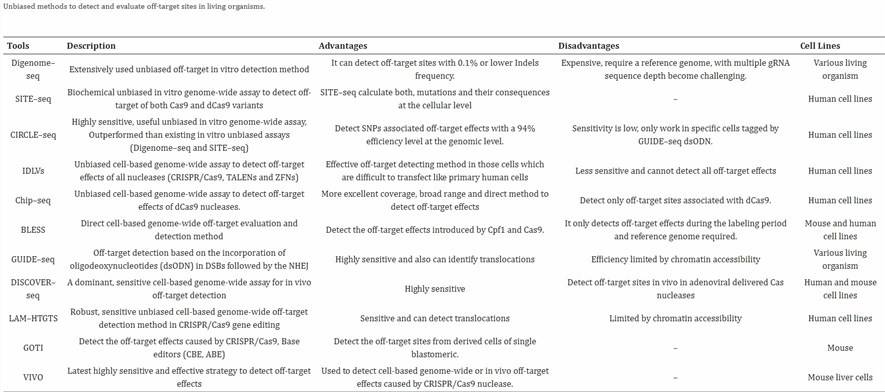
You may interested in
Learn More
Computational Simulation Methods of CRISPR-Cas9 off-target Detection
These techniques serve the purpose of predicting potential off-target sequences, primarily due to either the similarity between target sequences and other sequences or the limited specificity of guide RNAs (gRNAs). Potential off-target sequences can be deduced by integrating experimental data or employing computer algorithms. To validate off-target effects, one commonly resorts to PCR product sequencing. These approaches are often termed "biased," and the functioning of off-target prediction tools can be classified into two principal mechanisms: (1) models grounded in sequence alignment and (2) predictions relying on algorithm-based scoring [1].
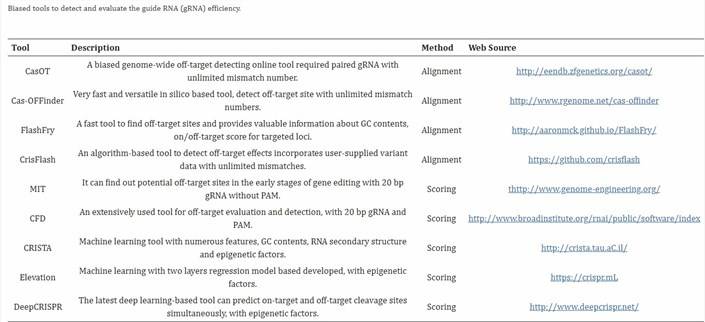
As the utilization of CRISPR technology becomes more widespread, the expectations regarding its application are also elevated. Often, when employing CRISPR technology, experimental designs may lack the inclusion of additional control groups, which could potentially pose challenges in subsequent experiments. Therefore, it is advisable to promptly perform off-target analysis after using CRISPR for gene editing or to retain early-stage samples. Both cells and animals are susceptible to spontaneous mutations.
References:
- Naeem, Muhammad et al. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells vol. 2020
- Ipek Tasan and Huimin Zhao. Targeting Specificity of the CRISPR/Cas9 System.ACS Synthetic Biology 2017
- Wienert, B., Wyman, S.K., Yeh, C.D. et al. CRISPR off-target detection with DISCOVER-seq. Nat Protoc 15, 1775–1799 (2020)


 Sample Submission Guidelines
Sample Submission Guidelines
