Next-Generation Sequencing in CRISPR Gene Editing Assessment and Quality Control
The advent of CRISPR-Cas9 gene editing technology has undeniably revolutionized the landscape of genetic research and therapeutic applications. Its ability to precisely modify specific regions of the genome has opened unprecedented opportunities for treating genetic disorders and driving advancements in biotechnology. However, harnessing the full potential of this revolutionary tool necessitates robust assessment and quality control measures to ensure the accuracy and safety of genomic manipulations. In this article, we delve into the cutting-edge role of Next-Generation Sequencing (NGS) in the comprehensive evaluation of CRISPR-edited genomes, exploring its capacity to detect on-target and off-target modifications, quantify editing efficiency, and its integration with advanced quality control techniques.
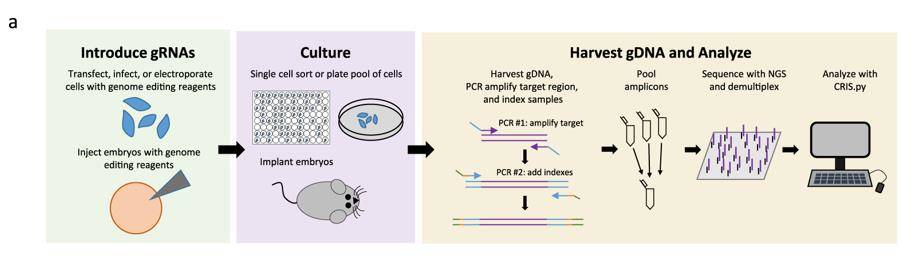 High-throughput Analysis for CRISPR-based Genome Editing. (Connelly et al., 2019)
High-throughput Analysis for CRISPR-based Genome Editing. (Connelly et al., 2019)
The Power of CRISPR-Cas9 Gene Editing
CRISPR-Cas9 is an RNA-guided endonuclease system derived from the prokaryotic immune system, enabling precise genome editing in various organisms. The Cas9 enzyme, guided by a single-guide RNA (sgRNA), targets specific DNA sequences, leading to double-stranded breaks. Subsequent cellular repair mechanisms, such as non-homologous end-joining (NHEJ) or homology-directed repair (HDR), result in gene knockout, insertion, correction, or targeted regulation of gene expression. This remarkable precision makes CRISPR-Cas9 a transformative technology with vast therapeutic potential and applications in functional genomics.
The Imperative of Accurate Assessment
While CRISPR-Cas9 technology offers remarkable precision, ensuring accurate and reliable gene editing outcomes is paramount. Even subtle errors or unintended modifications may compromise research findings and therapeutic developments. To address these challenges, a multidimensional approach to CRISPR gene editing assessment is required, encompassing in-depth analysis of on-target modifications, off-target effects, editing efficiency, and verification through stringent quality control measures.
NGS for Comprehensive CRISPR Gene Editing Assessment
- Detection of On-Target Modifications
NGS empowers researchers to interrogate the targeted genomic region at base-pair resolution, allowing for precise detection of on-target editing outcomes. Deep sequencing and variant calling analyses enable the identification of insertions, deletions, and precise modifications introduced by CRISPR-Cas9. To further enhance accuracy, amplicon-based sequencing and long-read sequencing technologies can be employed to resolve complex structural variations and allelic phasing, ensuring unambiguous annotation of edited loci.
- Identification of Off-Target Effects
The concern over off-target effects necessitates the application of sophisticated NGS-based strategies to uncover unintended genomic alterations. Utilizing whole-genome sequencing (WGS) or whole-exome sequencing (WES) approaches, researchers can comprehensively analyze the genome for potential off-target sites. Advanced computational algorithms, such as Digenome-seq enhance off-target profiling sensitivity, facilitating the detection of rare off-target events.
- Quantification of Editing Efficiency
Accurate quantification of editing efficiency is vital for optimizing CRISPR-Cas9 experiments and assessing the impact of various experimental parameters. NGS-based approaches enable allele quantification through amplicon sequencing or digital PCR, providing valuable insights into the percentage of edited alleles. Furthermore, targeted enrichment of editing-prone genomic regions using CRISPR-Cas9-based enrichment strategies combined with NGS allows for the fine-grained analysis of editing efficiency in specific genomic contexts.
Integration of Advanced Quality Control Techniques
- Library Preparation and Sequencing Quality
Rigorous quality control of NGS libraries is pivotal to minimize bias and experimental artifacts. Employing state-of-the-art library preparation kits and protocols ensures consistent and reproducible sequencing data. Additionally, incorporating molecular barcodes or molecular barcodes during library construction mitigates PCR amplification errors, further elevating data integrity.
- Bioinformatic Analysis
The deluge of data generated by NGS necessitates robust bioinformatic pipelines to process and interpret sequencing reads accurately. Implementing cutting-edge algorithms for alignment, variant calling, and annotation optimizes the identification and quantification of gene editing events. Furthermore, integrating single-cell sequencing approaches and spatial transcriptomics unravels the complexities of editing outcomes within heterogeneous cellular populations and tissues.
- Validation of Off-Target Sites
Thorough validation of potential off-target effects is indispensable for confirming NGS findings. Leveraging orthogonal validation methods, such as Sanger sequencing, digital droplet PCR (ddPCR), or genome-wide Cas9 activity assays, establishes the veracity of off-target events, solidifying research outcomes and refining the specificity of the CRISPR-Cas9 system.
Conclusion
CRISPR-Cas9 gene editing represents a paradigm shift in genomic research and therapeutic development. To harness the full potential of this transformative technology, comprehensive and accurate gene editing assessment is essential. Next-Generation Sequencing, with its capability to detect on-target and off-target modifications, quantify editing efficiency, and integrate advanced quality control techniques, serves as an indispensable tool in advancing precision and safety in CRISPR-Cas9 applications. By continuously refining NGS methodologies and embracing technological advancements, researchers pave the way for transformative gene editing therapies and discoveries that shape the future of medicine and biotechnology.
Reference:
- Connelly, Jon P., and Shondra M. Pruett-Miller. "CRIS. py: a versatile and high-throughput analysis program for CRISPR-based genome editing." Scientific reports 9.1 (2019): 4194.


 Sample Submission Guidelines
Sample Submission Guidelines
