RIP-Seq – Mapping the Binding Site of Protein-RNA Complex and Interpreting Epigenetic Regulation
Molecules inside the cell communicate with each other in order to execute the normal cell function. Biomolecules can form complexes with each other to perform a specific role. RNA and proteins are known to interact, forming complexes called ribonucleoproteins or RBPs that are important in post-transcriptional modification and regulation such as RNA cleavage, transport, localization, sequence editing, and other translational control. Analysis of the binding sites of these ribonucleoproteins is useful in understanding transcriptional and epigenetic regulation happening in the cell. Information received from these studies can be used to determine disease targets and development in the discovery of novel and effective drug targets.
Immunoprecipitation is used to isolate protein antigens by means of precipitation using the specificity of binding between particular protein antigens and their corresponding antibody. Results can then be analyzed in a microarray. This technique can also be used to precipitate proteins that are in contact with RNA (RIP-Chip). Another related technique is the RNA Immunoprecipitation Sequencing or RIP-Seq. Instead of analyzing the protein that bound to RNA, RNA is the target of RIP-Seq. The RNA component is sequenced using high-throughput sequencing techniques. RIP-Seq maps the binding site of the protein-RNA complex and provides the single-base sequence of the protein-bound RNA.
RIP-Seq is commonly used in studying epigenetic and transcriptomic controls of ribonucleoproteins. It is known to uncover the interaction of the two biomolecules at the genome-wide level. It was invented following the RIP-Chip technology developed by Keene et al. in 1999. Its relatively short and simple protocol does not require the use of complex laboratory equipment. The success of RIP-Seq analysis relies on the proper optimization of the immunoprecipitation step because it dictates the stringency of treatment in order to ensure that the RNA component is not damaged and the binding sites between the RNA and the protein are not destroyed.
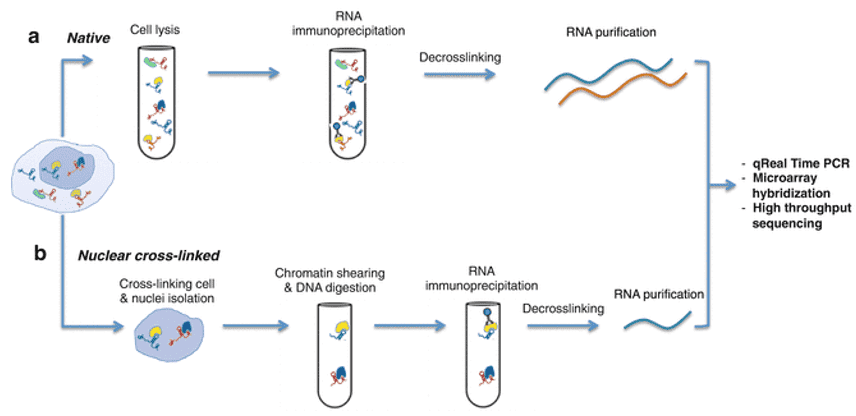
Figure 1. RNA immunoprecipitation sequencing protocol workflow. (Gagliardi, 2016)
During RNA Immunoprecipitation Sequencing, the target RNA-protein complex is precipitated with a corresponding antibody. RNase digestion is then conducted to remove extra RNA sequences that are not in contact with the protein. The resulting RNA is then extracted and reverse-transcribed to cDNA which is then sequenced and mapped back to the genome. This process can be used to study specific RNA-protein complexes with high resolution on the RNA sequence directly bound to the protein.
RIP-seq has been used in different studies to understand the cascade of activities inside the cell. Werfel, et al. used this technique to study the activities of microRNAs (miRNAs). They were able to demonstrate that the activity of mammalian miRNA is influenced by preferential binding to a particular 3' region, thus paving the way for the regulation between potential targets. Bersani, et al. used the technology to study the behavior of Wig-1, an RNA-binding protein that binds and regulates the expression of multiple mRNAs. Their study suggests novel binding sites for Wig-1 and a potential correlation between their results and stress response and tumor suppression.
RIP-seq technology has proven its usefulness in different studies relating to epigenetic and transcriptional control. It provides a view on the particular binding mechanisms of RNA-binding proteins which can be used to predict target mRNAs. It can also be used to study disease development including cancer and design novel therapeutics.
References:
- Bersani, C., et al. Genome-wide identification of Wig-1 mRNA targets by RIP-Seq analysis. Oncotarget, 2015, 7(2).
- Nicholson, C.O., Friedersdorf, M., Keene, J.D. Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. RNA Journal, 2016, 23(1).
- Wessels, H., Hirsekorn, A., Ohler, U. et al, N. Identifying RBP Targets with RIP-seq. Methods in Molecular Biology, 2016.
- Gagliardi M., Matarazzo M.R. RIP: RNA Immunoprecipitation. In: Lanzuolo C., Bodega B. (eds) Polycomb Group Proteins. Methods in Molecular Biology, 2016.


 Sample Submission Guidelines
Sample Submission Guidelines
