Ribosome Sequencing for Exploring Transcriptome And Translatome in Human Heart Disease Research
Translation is the process of ribosome reading mRNA template to guide protein synthesis, and is the key step of gene expression. Ribosome profiling (or Ribo-seq) is a deep sequencing-based tool that enables the detailed measurement of translation globally and in vivo, which is a study of the translation process from RNA to protein. This sequencing method provides the first systematic method for the annotation of the experimental coding region, thus discovering the gene expression regulation of a variety of complex biological processes, important aspects of protein synthesis mechanism, and even new proteins. The sequencing of these ribosomal protective fragments thus provides an accurate record of the ribosomal position at the time of translation. The distribution of ribosomal footprints can provide insights into translation control mechanisms (for example, it can be used to identify regulatory translation pauses and upstream open reading frameworks for translation). At present, such sequencing has been widely used in translation group research.
The strengths of Ribosomal sequencing
(1) It provides a large dynamic range of detection and quantification of translation in undisturbed cells.
(2) It provides uniquely rich and precise positional information.
(3) The instantaneous nature of the information collected reflects a snapshot of translation.
(4) Analytical advances that enable more comprehensive identification of other non-canonical translation events, will continue to expand our understanding of the protein-coding capacity of complex genomes.
(5) Sensitivity and resolution can measure the ribosomal behavior on specific transcripts in a single cell population, and the resolution can reach a single codon.
Ribo-seq for studying translatomics of hypertrophic cardiomyocytes
Hypertrophic growth of cardiomyocytes is one of the major compensatory responses in the heart after physiological or pathological stimulation. Enhanced protein synthesis mediated by messenger RNA translation is one of the main characteristics of cardiac hypertrophy. However, the mechanism of this induction at the translation level remains to be determined. Recently, researchers have analyzed the heart translation body by deep sequencing of Ribo-seq and RNA-seq. The data showed that the increase of protein synthesis efficiency in PE-induced cardiomyocyte hypertrophy was mainly due to the increase of ribosomal number, as evidenced by the increased translation of ribosomal protein genes. This study provides a complete genome overview of translation control behind cardiac hypertrophy through ribosome sequencing, and demonstrates the unrecognized role of micro-peptides in cardiac cell biology.
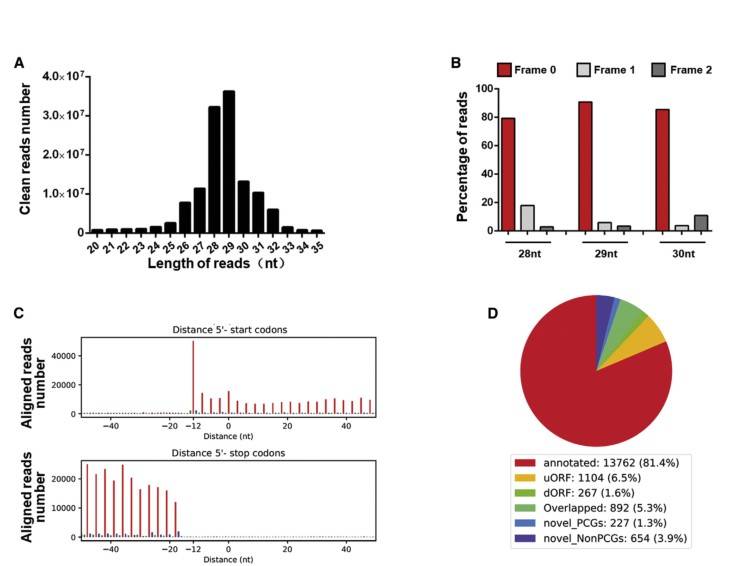 Detection of the active translating RNA fragments from hypertrophic cardiomyocytes
Detection of the active translating RNA fragments from hypertrophic cardiomyocytes
Ribosome profiling reveals the principles of translational control in human heart
Translation regulation is a key component of gene expression, but we know little about its role in human heart. Genome-wide translatomes can be characterized using ribosome profiling, which captures mRNA footprints protected by translating ribosomes. Researchers analyze the translatomes of 80 human hearts to identify new translation events and quantify the effect of translational regulation by mRNA-seq and ribo-seq. This study identified hundreds of previously undetected microproteins and suggest undiscovered roles of these microproteins or their involvement in biological functions assigned to the lncRNA. Additionally, 40 translatable circRNA molecules were found by analyzing the circRNA interface sequence captured by ribosomes in ribo-seq data. Among them, circCFLAR, circSLC8A1, circMYBPC3 and circRYR2 are the first translatable circRNA molecules found in myocardium.
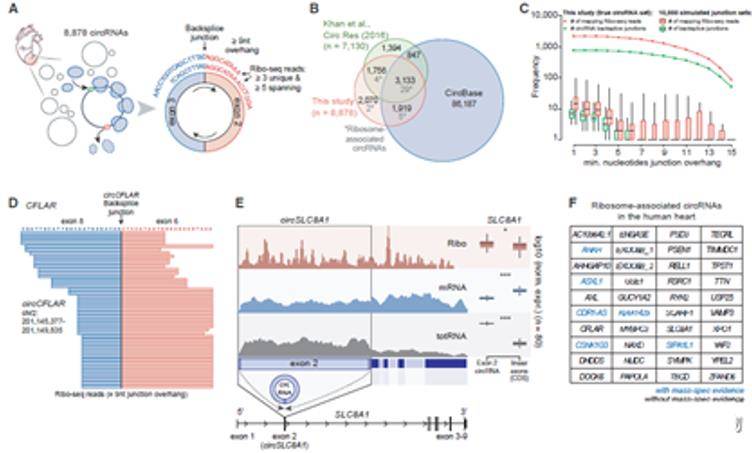 Ribo profiling analysis of translatable circRNA molecules
Ribo profiling analysis of translatable circRNA molecules
Ribosome profiling has rapidly become a widely used tool for understanding diverse and complex biological problems, which has a range of uses, from a broad proteomic tool to a specific probe of translation in an in vivo setting. The application of ribosome analysis in translation group can accurately and quantitatively describe what cells are translating, how translation is regulated, and the time and place of translation. The rich and quantitative nature of ribo-seq data provides an unprecedented opportunity to explore and simulate complex translation processes. We believe that ribosome profiling will provide valuable resources for in-depth understanding of human heart disease and its multiple potential mechanisms.
References:
- VanInsberghe, Michael et al. "Single-cell Ribo-seq reveals cell cycle-dependent translational pausing." Nature vol. 597,7877 (2021): 561-565.
- Brar, Gloria A, and Jonathan S Weissman. "Ribosome profiling reveals the what, when, where and how of protein synthesis." Nature reviews. Molecular cell biology vol. 16,11 (2015): 651-64.
- Yan, Youchen et al. "The cardiac translational landscape reveals that micropeptides are new players involved in cardiomyocyte hypertrophy." Molecular therapy: the journal of the American Society of Gene Therapy vol. 29,7 (2021): 2253-2267.
- van Heesch, Sebastiaan et al. "The Translational Landscape of the Human Heart." Cell vol. 178,1 (2019): 242-260.e29.


 Sample Submission Guidelines
Sample Submission Guidelines
