Enhancing Microbiome Function Prediction with 16s Full-Length Sequencing
The use of next-generation amplicon sequencing technology has sparked debates within the microbiome research community due to its inherent limitations, primarily its short read length and the subsequent scarcity of information it provides. The taxonomic precision of marker genes is a pivotal factor directly influencing the reliability and credibility of microbiome function prediction. Therefore, a key question arises: can function prediction based on next-generation amplicon sequencing results be considered credible, and is there a superior alternative to foster more efficient and precise research in the realm of function prediction?
A recent study delved deeply into the accuracy of function prediction and shed light on some critical concerns related to the biological soundness of microbiome function prediction. This study scrutinized the role of taxonomic composition, particularly when derived from short-read sequencing, and examined factors such as the taxonomic resolution of marker genes, the intragenomic variability of these genes, and the compositional nature of microbiome data. It convincingly posited that 16s full-length sequencing is poised to supplant short-read sequencing as the primary method for predicting microbiome function. This transition is expected to usher in a new era where 16s full-length sequencing will take center stage in microbiome function prediction, offering enhanced accuracy and credibility in this critical research domain.
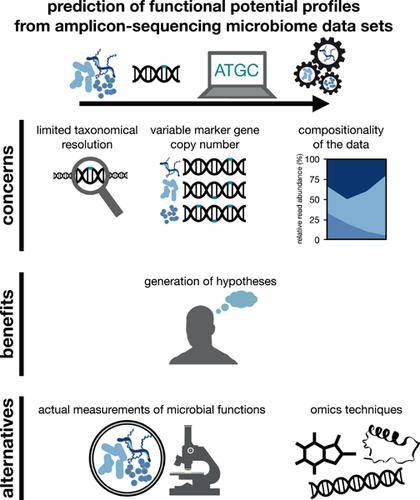 Predicting functional potential from microbiome taxonomic profiles generated by short-read amplicon sequencing. (Heidrich et al., 2022)
Predicting functional potential from microbiome taxonomic profiles generated by short-read amplicon sequencing. (Heidrich et al., 2022)
Taxonomic Resolution in 16s Full-Length Sequencing
In the realm of microbiology research, short read amplicon sequencing using the Illumina platform is the prevailing technology of choice. The PE300 sequencing mode, generating 2x300 base pair sequences, is widely adopted. In this domain, short-read, long amplicon sequencing is predominantly utilized to scrutinize the taxonomically informative segments of bacterial 16s ribosomal RNA (rRNA) and fungal internal transcribed spacer (ITS) loci. However, sequencing partial regions of these genes on the Illumina platform often exhibits limitations in species-level discrimination and struggles to differentiate closely related strains.
Recent advancements have showcased the potential of full-length 16s sequencing, achievable through long-read sequencing technologies like PacBio, to significantly enhance taxonomic resolution. 16s full-length sequencing comprehensively covers all nine variable regions of the 16S rRNA gene, thereby delivering a wealth of information surpassing that of single-region sequencing. It holds the promise of precise taxonomic identification at the species level and can effectively discern sequence polymorphisms while pinpointing taxonomic positions.
Furthermore, full-length 16S sequencing not only accurately identifies sequence polymorphisms but also allows for the localization of strain-level variations. This heightened sensitivity empowers the discovery of new microbial species and provides a more faithful representation of microbial community structures within samples.
While the cost of 16S full-length sequencing remains relatively high, it is expected that, in the long term, this technology, whether achieved through long-read amplicon sequencing or bioinformatics reconstruction from uniquely identified short-read amplicon sequencing, will gradually supplant short-read sequencing methods. This shift promises to elevate taxonomic resolution and, consequently, foster more accurate predictions of microbial functions.
Intragenomic Variability of Marker Genes
Another challenge in the analysis of microbiome structure using amplicon sequencing data lies in the varying copy numbers of marker genes within microbial genomes. This variability is occasionally misconstrued as allelic diversity, potentially introducing confusion into the microbiome composition analysis. Concurrently, intragenomic variability within marker genes hinders the accurate assessment of the relative importance of potential functions, possibly leading to an overestimation of the diversity within potential functional profiles.
The limitations of Next-Generation Sequencing (NGS) technology become evident in its inability to precisely identify intragenomic genetic variability. This is primarily due to the technology's short read length and restricted sequencing coverage. In stark contrast, 16S full-length sequencing, characterized by its superior coverage and accuracy, excels in accurately identifying sequence polymorphisms and pinpointing strain-level variations.
The Compositional Nature of Microbiome Data
One critical aspect to consider is that the number of sequence reads generated by Next-Generation Sequencing (NGS) from a sample does not directly correlate with the number of bacterial cells within that sample, making it problematic to translate reads into bacterial abundance. NGS reveals only the relative size of a specific segment of the microbial community represented by each taxonomic unit. This characteristic assembles the NGS microbiome dataset to provide insights into the relative abundance of sequencing reads, expressed as proportions or frequencies of taxa in a microbial community. However, it falls short in providing information on the absolute abundance of taxa, primarily because the overall size (microbial biomass) of the community remains undisclosed.
Consequently, even if the predicted functional profile of a community aligns with its actual function, estimating the magnitude of the functional potential becomes a formidable challenge due to the lack of consideration for the total population size of the community. Various microbial quantification methods exist to address this limitation, but they can introduce additional data heterogeneity.
One alternative approach to mitigate the issue of analyzing microbial compositional data is the use of ratios. This method eliminates bias stemming from microbial self-loading and helps overcome challenges associated with PCR amplification bias, a well-recognized source of error that can distort community composition and functional predictions.
Uncovering the Genuine Function of the Microbiota
Another more intricate approach involves the measurement of the actual function of the microbiota whenever feasible. However, this endeavor is only attainable under specific conditions: the microbial community must be accessible, the target function must be actively engaged at the time of sampling, and there should be a sufficiently large population size and sample volume available for analysis. For microbiota that does not meet these criteria, additional techniques such as real-time fluorescent quantitative PCR serve as valuable complements for quantitatively characterizing the genetic potential of certain functional genes. It's vital to recognize that these genetic potentials do not necessarily equate to actual microbial activities and processes.
From this perspective, histological techniques such as transcriptomics, proteomics, and metabolomics prove instrumental in exploring the genetic potential that has been translated into action. Combining functional prediction with the measurement of specific functions represents a potent approach for gaining insights into microbial functionality.
Reference:
- Heidrich, Vitor, and Lukas Beule. "Are short‐read amplicons suitable for the prediction of microbiome functional potential? A critical perspective." iMeta 1.3 (2022): e38.


 Sample Submission Guidelines
Sample Submission Guidelines
