What is Cycle Sequencing
In the domain of genetic analysis, cycle sequencing emerges as a pinnacle of precision and efficacy, providing researchers with a potent instrument for unraveling the complexities embedded within DNA. Derived as an enhancement of the conventional Sanger sequencing technique, cycle sequencing has metamorphosed the genetic sequencing panorama, facilitating exhaustive analysis characterized by unprecedented precision and celerity. Within this discourse, we embark on a comprehensive exploration of cycle sequencing, elucidating its foundational tenets, procedural nuances, and its myriad applications within the sphere of genetic scrutiny.
Understanding Cycle Sequencing: A Closer Look at the Core Principles
Cycle sequencing epitomizes a nuanced fusion of molecular biology methodologies, harmonizing facets of classical Sanger sequencing with the efficacy inherent in polymerase chain reaction (PCR). At its core, cycle sequencing endeavors to elucidate the genetic blueprint encapsulated within DNA molecules with an unparalleled degree of precision and fidelity.
Principle of Chain Termination
At the heart of cycle sequencing lies the pivotal principle of chain termination, a foundational aspect of the Sanger sequencing paradigm. Within this framework, DNA synthesis encounters interruption at precise intervals along the DNA template through the integration of dideoxynucleotides (ddNTPs). Devoid of a 3'-OH group, these ddNTPs impede the subsequent addition of nucleotides by the DNA polymerase enzyme, thereby inducing the cessation of DNA synthesis.
Integration of PCR Cycling
The distinctive feature that distinguishes cycle sequencing from its traditional counterpart, Sanger sequencing, is its incorporation of PCR-like cycling. In contrast to the linear trajectory of DNA synthesis observed in Sanger sequencing, cycle sequencing leverages thermal cycling within a thermal cycler to orchestrate iterative denaturation, annealing, and extension of DNA fragments. This cyclic modality amplifies the efficiency and resilience of the sequencing reaction, thereby empowering the acquisition of sequencing data even from scant amounts of template DNA.
Utilization of Thermo-Stable DNA Polymerase
At the crux of cycle sequencing lies a pivotal facilitator: the employment of a thermo-stable DNA polymerase, exemplified by Taq polymerase. This enzymatic entity manifests exceptional resilience under elevated temperatures, thereby facilitating the iterative denaturation and synthesis cycles intrinsic to cycle sequencing. Furthermore, the thermo-stability inherent to the DNA polymerase mitigates the probability of enzyme denaturation, thus safeguarding the fidelity of DNA synthesis across the entirety of the sequencing endeavor.
Labeling of Sequencing Primers
Yet another crucial facet of cycle sequencing pertains to the labeling of sequencing primers, pivotal entities that initiate DNA synthesis. Conventionally, these primers are tagged with either fluorescent dyes or radioactive isotopes, facilitating the visualization and detection of the synthesized DNA fragments. Through the integration of labeled primers into the sequencing milieu, researchers can meticulously decipher the nucleotide sequence embedded within the DNA template.
Fragment Separation and Analysis
After the culmination of the cycle sequencing procedure, the synthesized DNA fragments undergo segregation according to size, employing either capillary electrophoresis or gel electrophoresis. Subsequently, the fluorescently labeled fragments are identified and scrutinized, wherein each fluorescent signal corresponds to a distinct nucleotide assimilated during DNA synthesis. Leveraging intricate data analysis algorithms, researchers undertake the intricate task of deciphering the DNA sequence by deducing the sequential arrangement of fluorescent signals detected.
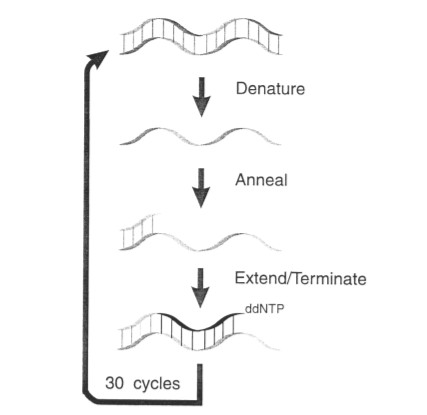 Figure 1 Outline of the cycle sequencing reaction. Double- or single-stranded templates are cycled through a three-temperature cycling reaction consisting of high-temperature denaturation, primer annealing, and primer extension/termination (Keith Kretz,et al,.2024)
Figure 1 Outline of the cycle sequencing reaction. Double- or single-stranded templates are cycled through a three-temperature cycling reaction consisting of high-temperature denaturation, primer annealing, and primer extension/termination (Keith Kretz,et al,.2024)
Cycle Sequencing vs PCR
Both cycle sequencing and PCR are molecular biology techniques employed in DNA analysis, yet they serve divergent purposes and utilize distinct methodologies. PCR primarily serves to amplify specific DNA sequences, thereby facilitating the generation of numerous copies of a targeted DNA region. Conversely, cycle sequencing is directed towards elucidating the nucleotide sequence of a DNA fragment.
| PCR | Cycle Sequencing | |
| Methodology | PCR involves iterative cycles of denaturation, annealing, and extension, culminating in exponential amplification of the targeted DNA region. | Cycle sequencing adopts PCR-like thermal cycling to enable repetitive denaturation, primer annealing, and DNA synthesis. However, its primary aim is DNA sequencing rather than amplification. |
| Enzymes Used | PCR typically relies on DNA polymerase enzymes, such as Taq polymerase, tailored for DNA amplification. | In cycle sequencing, a thermo-stable DNA polymerase is also employed. Nevertheless, its function is to synthesize DNA fragments during the sequencing process, diverging from the amplification role observed in PCR. |
| Purpose | PCR serves to amplify specific DNA sequences, facilitating various applications such as cloning, genotyping, and mutation detection. | Cycle sequencing is dedicated to elucidating the nucleotide sequence of DNA fragments, thereby enabling the unraveling of genetic information embedded within the DNA. |
Cycle Sequencing vs Sanger Sequencing
Cycle sequencing and Sanger sequencing, while both belonging to the realm of DNA sequencing methods, exhibit distinctions in their methodologies and approaches. Sanger sequencing, conceived by Frederick Sanger in the 1970s, represents the conventional modality of DNA sequencing, whereas cycle sequencing emerges as an evolved iteration of this technique, devised to augment efficiency and throughput.
| Sanger Sequencing | Cycle Sequencing | |
| Methodology | Sanger sequencing entails the introduction of chain-terminating dideoxynucleotides (ddNTPs) into DNA strands, resulting in the generation of a spectrum of terminated fragments with varying lengths. | Cycle sequencing adopts the utilization of chain-terminating ddNTPs while integrating PCR-like thermal cycling to facilitate iterative denaturation, annealing, and extension processes, thereby enhancing the efficacy of the sequencing endeavor. |
| Efficiency and Throughput | Sanger sequencing is constrained by its labor-intensive nature and limited throughput, necessitating individual execution of each sequencing reaction. | Conversely, cycle sequencing boasts heightened efficiency and throughput, enabled by the capacity for simultaneous execution of multiple sequencing reactions within a thermal cycler, thereby curtailing the time and labor investment in sequencing operations. |
| Application | Sanger sequencing finds utility in sequencing discrete DNA fragments, such as plasmids or PCR products. | In contrast, cycle sequencing finds optimal application in high-throughput sequencing scenarios, including the sequencing of entire genomes or expansive DNA libraries, owing to its amplified efficiency and scalability. |
You may interested in
Learn More
The Steps of Cycle Sequencing
Cycle sequencing, an extension of the classical Sanger sequencing method, encompasses a series of pivotal procedures aimed at the precise amplification and sequencing of DNA fragments.
1. Primer Labeling and Preparation
The initial step entails the labeling of sequencing primers, meticulously designed to anneal to the target DNA sequence, thereby instigating DNA synthesis. Within cycle sequencing, the primers are imbued with either a fluorescent dye or a radioactive label, facilitating subsequent detection. These labeled primers undergo meticulous preparation, wherein they are annealed to the single-stranded template DNA, concomitant with the presence of a DNA polymerase enzyme and a blend of deoxynucleotides (dNTPs).
2. Thermal Cycling
Following the primer annealing phase within the template DNA, the reaction mixture is subjected to a process known as thermal cycling. This crucial process encompasses a regimented series of temperature oscillations designed to expedite the denaturation of DNA, facilitate primer annealing, and stimulate DNA synthesis.
Typically, the reaction mixture is first exposed to an elevated thermal condition conducive to DNA denaturation. This calculated rise in temperature induces the separation of the double-stranded DNA into individual strands. Subsequently, a temperature reduction is instigated, promoting the primer annealing phase. During this latter step, the primers selectively bind to their respective complementary sequences present within the template DNA.
Lastly, an intermediate temperature regime is employed to initiate and foster the DNA synthesis process. Throughout this phase, the enzymatic machinery represented by the DNA polymerase substantively extends the primer. This extension process is achieved through the polymerase's inherent ability to sequentially incorporate complementary nucleotides into the template strand, thus guiding the proper architectural assembly of the new DNA molecule.
3. Incorporation of Chain-Terminating Nucleotides
Upon the commencement of the DNA synthesis phase, a species of nucleotides known as dideoxynucleotides (ddNTPs), referred to as chain-terminating nucleotides, are assimilated into the developing DNA filament. These particular ddNTPs are devoid of a 3'-OH group, an aspect which precludes any subsequent continuation of DNA synthesis upon their addition to the expanding strand. Consequently, this prompts the cessation of DNA synthesis at each respective ddNTP location, culminating in the production of an array of DNA fragments of disparate lengths.
4. Separation and Detection
Upon the culmination of the thermocyclic reactions, the generated DNA fragments are distinguished according to their respective sizes via modalities such as gel electrophoresis or capillary electrophoresis. Following separation, these fragments undergo detection and subsequent visualisation either through fluorescence detection, should the primers carry a fluorescent label, or by means of autoradiography if the primers have been labelled radioactively. The resultant sequence data can then be scrutinised for ascertaining the nucleotide sequence of the original DNA template.
By adhering to these procedural steps, cycle sequencing facilitates the precise and efficient sequencing of DNA fragments. This renders it an invaluable technological asset for molecular biology research, diagnostic practices, and an array of other scientific disciplines.
Cycle Sequencing Protocol: A Detailed Procedure
Materials and Equipment
Before starting the cycle sequencing protocol, gather all necessary materials and equipment:
Materials:
Labeling buffer: 300 mM Tris-HCl, pH 7.8, 50 mM MgCl2, 1 M KCl.
Kinase dilution buffer: 50 mM Tris-HCl, pH 7.6, 25 mM KCl, 5 mM DTT, 0.1 mM ATP, 0.2 mg/mL BSA, 50% glycerol.
Sequencing primer: Commercially available, e.g., 5'-CCCAGTCACGACGTTGTAAAACG-3'.
Labeled adenosine triphosphate: [γ-32P]ATP (≥ 3000 Ci/mmol, 10 mCi/mL) or [γ-33P]ATP (1600 Ci/mmol).
Sequencing buffer: 300 mM Tris-HCl, pH 9.0, 50 mM MgCl2, 300 mM KCl, 0.5% W-1.
Formamide dye solution: 95% deionized formamide, 10 mM EDTA, pH 8.0, 0.1% bromophenol blue, 0.1% xylene cyanol.
Taq DNA polymerase, dNTPs, and ddNTPs: Commercially available.
10X TE buffer: 100 mM Tris-HCl, pH 7.5, 1 mM EDTA.
Light silicone oil: Optional.
PCR primers with universal tails: e.g., M13 forward (5'-TGTAAAACGACGGCCAGT-3') and M13 reverse (5'-CAGGAAACAGCTATGACC-3').
Equipment:
Thermal cycler: Capable of executing PCR-like thermal cycling.
Water baths or heater blocks: For maintaining specific temperatures.
Microcentrifuge tubes: Polypropylene, 0.2 or 0.5 mL.
Automatic pipettes: Capable of dispensing 0.5–20 µL and 10–100 µL.
Electrophoresis equipment: For DNA sequencing.
X-ray film: Single-sided with a blue base.
Procedure
Step 1: Primer Labeling
Dilute the sequencing primer to 0.5 pmol/µL with 1X TE buffer.
Mix 2 µL of the diluted primer, 1 µL of labeling buffer, 2 pmol of labeled ATP, and 1 µL of T4 polynucleotide kinase in a microcentrifuge tube.
Incubate the reaction at 37°C for 10 minutes, followed by 55°C for 5 minutes.
Place the tube on wet ice to terminate the reaction.
Step 2: Preparing Reaction Mixtures
Prepare a prereaction mix by combining 5 µL of end-labeled primer, 4.5 µL of sequencing buffer, 7–26 µL of template DNA, 0.5 µL of Taq DNA polymerase, and autoclaved distilled water to a total volume of 36 µL.
Divide the prereaction mix into four microcentrifuge tubes labeled A, C, G, and T.
Step 3: Sequencing Reactions
Prepare sequencing mixes according to Table 1.
Add 10 µL of the appropriate sequencing mix to each labeled microcentrifuge tube.
Securely cap the tubes and mix gently.
If using a thermal cycler without a heated lid, add 20 µL of silicone oil to each reaction.
Initiate thermal cycling with the following program:
3 min at 95°C for denaturation.
20 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 70°C for 60 s.
10 additional cycles of denaturation at 95°C for 30 s and extension at 70°C for 60 s.
Step 4: Analysis
Terminate the reactions by adding 5 µL of formamide dye solution to each tube.
Mix well and centrifuge briefly.
Store the reaction tubes at -20°C until analysis.
Separate the reaction products on a sequencing gel and visualize using X-ray film.
Applications of Cycle Sequencing
Genetic Disease Research
Cycle sequencing is pivotal in genetic disease research as it facilitates the precise determination of DNA sequences linked to hereditary disorders. For instance, Ng et al. (2010) employed cycle sequencing to unearth novel mutations within the PANK2 gene, implicated in pantothenate kinase-associated neurodegeneration (PKAN), a rare neurodegenerative ailment. Through sequencing the coding segments of the PANK2 gene in afflicted subjects, the investigators pinpointed missense mutations that impeded protein functionality, thereby enhancing comprehension of the genetic underpinnings of PKAN (Ng et al., 2010).
Environmental Microbiology
In the realm of environmental microbiology, cycle sequencing serves as a cornerstone method for delineating microbial communities and their repertoire of functional genes within diverse ecosystems. For instance, Liu et al. (2018) harnessed cycle sequencing of 16S rRNA genes to scrutinize microbial diversity and composition within soil samples sourced from various land-use categories. Through the sequencing of PCR-amplified fragments of 16S rRNA genes, the researchers discerned alterations in microbial community architecture across distinct land-use patterns, thereby illuminating the repercussions of land management practices on soil microbial assemblages (Liu et al., 2018).
Cancer Genomics
The technique of cycle sequencing finds utility in the expansive field of cancer genomics, where it is employed to unearth somatic mutations and associated genomic alterations pivotal to tumorigenesis and subsequent progression. This is eloquently highlighted in the seminal study executed by Imielinski et al. (2012), where the concerted application of cycle sequencing unraveled the mutational panorama of lung adenocarcinoma. Through a methodical sequencing of tumor and corresponding normal DNA samples, the investigators successfully pinpointed recurrent mutations in certain genes, namely EGFR, KRAS, and TP53. This revelation supplied important clues pertaining to the molecular mechanisms that influence the pathogenesis of lung cancer (Imielinski et al., 2012).
Forensic Analysis
In forensic science, cycle sequencing stands as a fundamental technique for DNA profiling and forensic analysis, enabling the identification of individuals through their genetic signatures. For example, Budowle et al. (2003) utilized cycle sequencing to scrutinize short tandem repeat (STR) loci for forensic DNA profiling. By sequencing PCR-amplified fragments of STRs extracted from both crime scene evidences and reference samples, the researchers successfully matched DNA profiles, thus contributing significantly to criminal inquiries and the dispensation of justice (Budowle et al., 2003).
The utility of cycle sequencing spans across diverse domains, encapsulating a spectrum of biological research endeavors and real-world applications. From unraveling the genetic underpinnings of diseases to delineating microbial communities and profiling forensic DNA, cycle sequencing emerges as a versatile instrument for generating precise DNA sequence information. Its multifaceted applications contribute significantly to the advancement of knowledge in genetics, microbiology, oncology, and forensic science, thereby enriching our comprehension of these intricate scientific realms.
Expanding Service Offerings: Incorporating Sanger and Next Generation Sequencing
In exploring the intricacies of cycle sequencing's contribution to genetic analysis, it is imperative to contextualize its significance within the landscape of sequencing methodologies. Although cycle sequencing serves as a fundamental method, offering meticulous and expedient DNA analysis, its efficacy is augmented and harmonized by the functionalities inherent in both Sanger sequencing and Next Generation Sequencing (NGS).
Sanger Sequencing: A Foundation of Precision
Sanger sequencing, credited to the pioneering work of Frederick Sanger, endures as the quintessential method in DNA sequencing owing to its meticulous elucidation of nucleotide sequences. By employing the strategic termination of DNA synthesis utilizing chain-terminating ddNTPs, Sanger sequencing empowers researchers to discern genetic sequences with unrivaled precision. Its focused methodology proves especially advantageous in the validation of genetic variants and the sequencing of designated regions with utmost fidelity.
Next Generation Sequencing: Revolutionizing Genetic Analysis
At the forefront of high-throughput genomics, NGS technologies have revolutionized our methodology in genetic analysis. Through the parallelization of sequencing reactions and the utilization of pioneering platforms, NGS facilitates swift and exhaustive genomic elucidation on a magnitude previously inconceivable with conventional sequencing techniques. Spanning from whole genome sequencing to metagenomics and transcriptomics, NGS empowers researchers to delve into the intricacies of the genome with unparalleled depth and scope.
Integration into Service Offerings
In concert with our commitment to thorough investigation through cycle sequencing, our facility now broadens its scope to include Sanger sequencing and NGS. This holistic strategy guarantees researchers access to a varied repertoire of sequencing methodologies, each endowed with distinctive capabilities and utilities. Whether unraveling the genetic underpinnings of diseases, characterizing microbial populations, or profiling forensic DNA, our enhanced service portfolio equips researchers with the requisite tools and proficiency to adeptly navigate the complexities of genetic analysis.
References:
- Ng, J., Papandreou, A., Heales, S. J., Kurian, M. A. (2010). Monoamine neurotransmitter disorders—Clinical advances and future perspectives. Nature Reviews Neurology, 6(12), 705-715.
- Liu, J., Sui, Y., Yu, Z., Shi, Y., Chu, H., Jin, J., ... & Wang, G. (2018). Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biology and Biochemistry, 121, 103-108.
- Imielinski, M., Berger, A. H., Hammerman, P. S., Hernandez, B., Pugh, T. J., Hodis, E., ... & Garraway, L. A. (2012). Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell, 150(6), 1107-1120.
- Budowle, B., Moretti, T. R., Baumstark, A. L., Defenbaugh, D. A., Keys, K. M., & Brown, A. L. (2003). Validation of short tandem repeats (STRs) for forensic usage: Performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. Journal of Forensic Sciences, 48(4), 928-956.
- Blakesley, R.W. (2001). Cycle Sequencing. In: Graham, C.A., Hill, A.J.M. (eds) DNA Sequencing Protocols. Methods in Molecular Biology
- Surdhar, G.K. (2002). Cycle Sequencing of PCR Products. In: Theophilus, B.D.M., Rapley, R. (eds) PCR Mutation Detection Protocols. Methods in Molecular Biology, vol 187. Humana Press.


 Sample Submission Guidelines
Sample Submission Guidelines
