Advancing Plant Disease Resistance Research through Single Cell Sequencing
Plant diseases pose a significant threat to global agriculture, causing substantial yield losses and economic repercussions. Traditional approaches to studying plant disease resistance have provided valuable insights but are limited in capturing the dynamic cellular and molecular responses during infection. In recent years, single-cell sequencing has emerged as a transformative technology, enabling researchers to dissect the heterogeneity of plant tissues and unravel the intricate mechanisms of disease resistance at the cellular level.
Plant Diseases on Global Agriculture
Plant diseases caused by pathogens like fungi, bacteria, viruses, and nematodes have a significant impact on global agriculture. They result in substantial crop losses, threatening food security, farmers' livelihoods, and the global economy. These diseases reduce crop yields, disrupt supply chains, and increase prices, leading to economic losses. They also pose a threat to food security, particularly in vulnerable regions. Plant diseases can trigger trade barriers, affect market access, and necessitate the use of pesticides with environmental and health consequences. They disrupt ecosystems, reduce biodiversity, and require a comprehensive approach involving prevention, early detection, resilient crop varieties, and international collaboration for effective management.
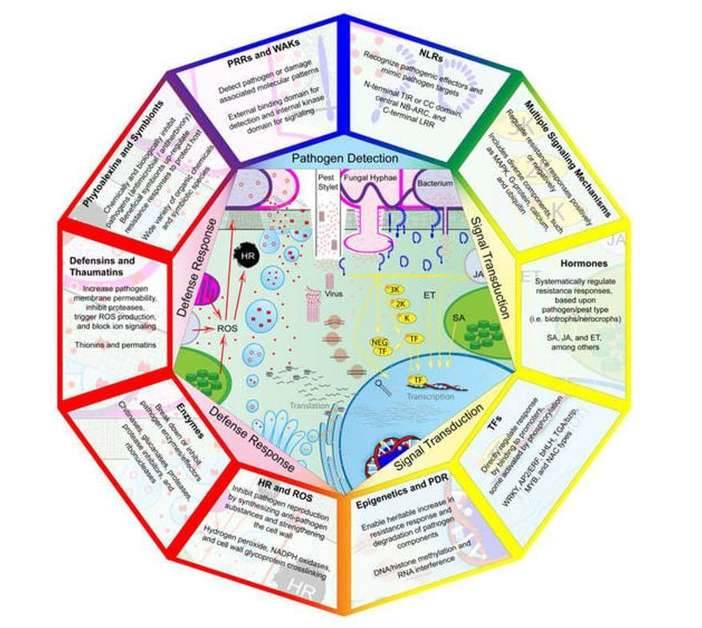 Components of plant disease resistance mechanisms involved in pathogen detection, signal transduction, and defense response. (Andersen et al., 2018)
Components of plant disease resistance mechanisms involved in pathogen detection, signal transduction, and defense response. (Andersen et al., 2018)
Traditional Approaches and Limitations in Plant Disease Resistance Research
Understanding the mechanisms underlying plant disease resistance is crucial for developing effective strategies to combat plant pathogens. Traditional approaches, including breeding for disease resistance and phenotypic characterization, have contributed to the development of resistant plant varieties. However, these methods have limitations in capturing the complex cellular and molecular dynamics during infection.
- Breeding for disease resistance: Challenges and time-consuming processes
Breeding programs traditionally rely on phenotypic evaluations and selection based on disease resistance. However, this approach is time-consuming, as it requires multiple generations of crosses and phenotypic assessments. Moreover, it may overlook crucial molecular mechanisms underlying resistance.
- Phenotypic characterization: Incomplete understanding of molecular mechanisms
Phenotypic characterization provides valuable information about disease resistance traits but often lacks the molecular resolution required to fully comprehend the complex regulatory networks involved. It is challenging to capture the dynamic changes in gene expression and cellular responses that occur during infection.
- Need for a comprehensive approach to uncover cellular heterogeneity
Plant tissues consist of diverse cell types, each playing a distinct role in disease resistance. Traditional approaches, which analyze bulk tissue samples, mask the heterogeneity within tissues, preventing a comprehensive understanding of cell-specific responses to pathogens. This knowledge gap hampers the development of targeted interventions."
Single Cell Sequencing in Plant Research
Single-cell sequencing involves isolating and sequencing the genetic material (DNA or RNA) from individual cells, allowing researchers to characterize the transcriptomic or genomic profiles of each cell. Various technologies, such as droplet-based methods and microwell platforms, enable high-throughput analysis of thousands of individual cells in a single experiment.
Single-cell sequencing is a cutting-edge technology that enables the high-resolution analysis of individual cells, providing insights into their genetic and transcriptional profiles. In the context of plant disease resistance research, single-cell sequencing offers unprecedented opportunities to decipher the heterogeneity within plant tissues, unravel cellular responses to pathogen attack, and identify novel genes and pathways involved in disease resistance.
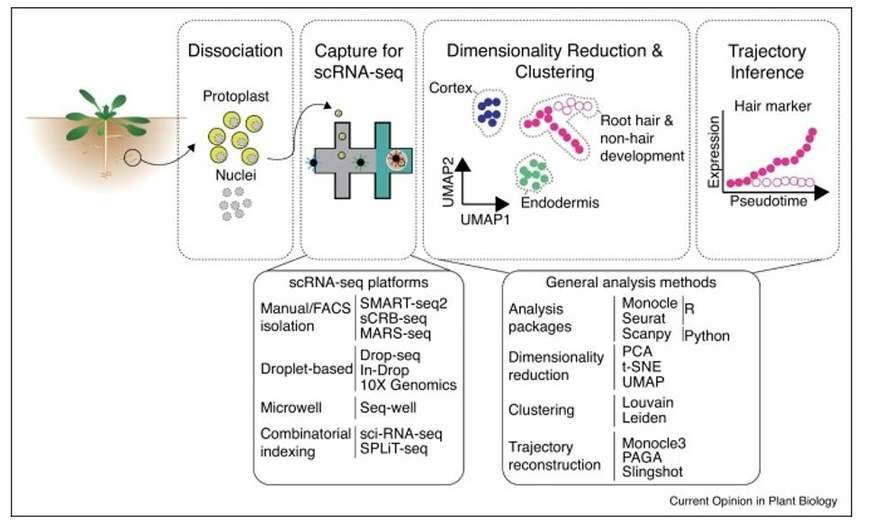 Single-cell genomics in plants. (McFaline-Figueroa et al., 2020)
Single-cell genomics in plants. (McFaline-Figueroa et al., 2020)
First of all, single-cell sequencing allows for the identification of crucial molecular mechanisms underlying disease resistance. By sequencing individual cells, researchers can analyze gene expression patterns, identify specific cell types or subpopulations involved in resistance, and uncover novel regulatory networks. This approach provides a more detailed and comprehensive understanding of the molecular basis of disease resistance, enabling targeted breeding programs and the development of resistant crop varieties in a shorter time frame.
As for phenotypic characterization, single-cell sequencing provides a higher-resolution view of the molecular changes that occur during infection. By capturing gene expression profiles at the single-cell level, researchers can observe dynamic changes in gene expression and cellular responses in real-time. This detailed information helps in deciphering the complex regulatory networks involved in disease resistance and provides insights into the underlying mechanisms that drive resistance or susceptibility.
Single-cell sequencing enables the characterization of cellular heterogeneity within plant tissues. By analyzing individual cells, researchers can identify and classify different cell types and subpopulations based on their gene expression profiles. This comprehensive approach uncovers the specific responses of different cell types to pathogens, revealing the heterogeneity within tissues that would be masked in traditional bulk tissue analyses. Understanding cell-specific responses is crucial for developing targeted interventions and breeding strategies to enhance disease resistance.
References:
- Andersen, Ethan J., et al. "Disease resistance mechanisms in plants." Genes 9.7 (2018): 339.
- McFaline-Figueroa, José L., Cole Trapnell, and Josh T. Cuperus. "The promise of single-cell genomics in plants." Current opinion in plant biology 54 (2020): 114-121.


 Sample Submission Guidelines
Sample Submission Guidelines
