Utilizing RNA Sequencing for The Analysis of Host-Microbe Interactions
Within the intricate interplay between host cells and commensal/pathogenic bacteria, the cellular milieu plays a pivotal role in determining the eventual outcome of these interactions. This includes the activation or suppression of bacterial virulence programs, as well as the engagement of host defense mechanisms or tolerance responses. To gain a comprehensive understanding of the fundamental principles underlying host-microbe interactions in both healthy and diseased states, we require high-resolution methodologies capable of capturing their intricate dynamics across various scales. This encompasses investigations spanning from the broader context of the whole organism down to the nuanced intricacies of single-cell interactions, all of which contribute to establishing a robust theoretical foundation for the management of microbe-associated diseases.
Three Evolutionary Phases of Transcriptomics in Host-Microbe Interaction Studies
In the realm of infection biology, researchers employ various levels of assays to delve into host-microbe interactions, such as genomics, epigenomics, transcriptomics, proteomics, metabolomics, and more. Among these, transcriptomics has gained significant traction for its ability to offer a snapshot of cellular physiology, owing to its sensitivity, cost-effectiveness, and widespread applicability. Since the inception of transcriptome sequencing in the field of infection biology roughly a decade ago, we have witnessed the emergence of three distinct phases in the analysis of host-microbe interactions.
In the initial phase (Single-Sided RNA Sequencing/One-Sided Study), host cells and microbes are physically separated, with an emphasis on examining either the host or the microbe exclusively.
The second phase marks a pivotal shift where both sides of the interaction are examined simultaneously. This phase encompasses the profiling of host and microbe interactions (Dual RNA-Seq or dual expression profiling) and delves into metatranscriptomics to explore the broader microbiome landscape. Concurrently, there has been a reevaluation of the role of non-coding transcripts, which were previously considered mere by-products of RNA-seq analysis. These non-coding transcripts have proven to be invaluable in uncovering crucial insights into host-microbe interactions.
In the third and current phase, we witness the ascendancy of single-cell bacterial transcriptome sequencing, a technique increasingly employed to unravel the heterogeneity inherent in host-microbe interactions. This phase represents a powerful advancement in our ability to scrutinize the intricate dynamics at the single-cell level within the microbial component of these interactions.
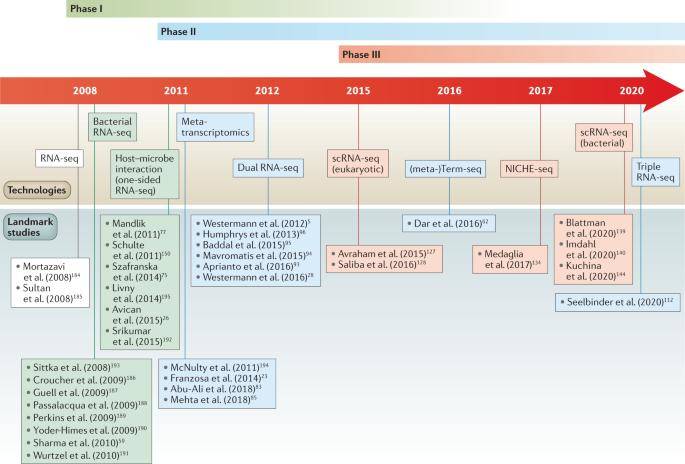 The history of RNA-seq-based infection research. (Westermann et al., 2021)
The history of RNA-seq-based infection research. (Westermann et al., 2021)
Detection Methods for Host-Microbe Interactions
- Investigate microbial community structure and function through metatranscriptome sequencing.
- Employ dual RNA-seq to analyze both pathogen and infected host tissue.
- Conduct triple RNA-seq to dissect viral and bacterial coinfections or competing symbionts with their hosts.
- Utilize single-cell transcriptome sequencing for host analysis or single bacterial transcriptome sequencing.
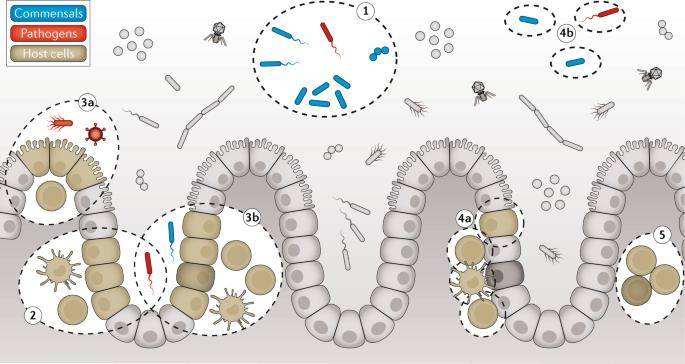 Graphical overview of RNA-seq-based approaches to study inter-species interactions in the mammalian intestine. (Westermann et al., 2021)
Graphical overview of RNA-seq-based approaches to study inter-species interactions in the mammalian intestine. (Westermann et al., 2021)
Metatranscriptome Sequencing
Metatranscriptome sequencing is employed to delve into the intricacies of microbial community structure and functionality. Achieving precise quantification of gene expression within the microbiome necessitates a meticulous process. To simplify, metagenomic and metatranscriptomic data from the same sample are initially processed independently. Subsequently, metagenomic data is utilized to normalize metatranscriptomic data, yielding MT/MG values that encapsulate gene, species, and functional activity changes.
Notably, studies on the gut microbiome have uncovered a greater degree of variability in the metatranscriptome compared to the metagenome. This observation implies that microbes exhibit active regulation in response to shifts in their environment and nutritional conditions. The shifts in both composition and functionality of human gut microbes have been linked to disease infections. For instance, the abundance of gut microbial mRNAs encoding proteins involved in antioxidant defense and metal ion homeostasis has been identified as a negative correlate with the risk of experiencing typhoid fever symptoms following infection with Salmonella typhimurium.
Dual RNA-seq
Dual RNA-seq, an innovative approach, is employed in the investigation of interactions between pathogens and mammalian hosts at the transcriptomic level. This cutting-edge technique enables the concurrent analysis of gene expression within bacterial organisms and infected host cells or tissues, whether intracellular or extracellular. Through dual RNA-seq, we aim to unveil previously undiscovered intricacies within host-microbe interactions, promising a deeper understanding of these interdependencies.
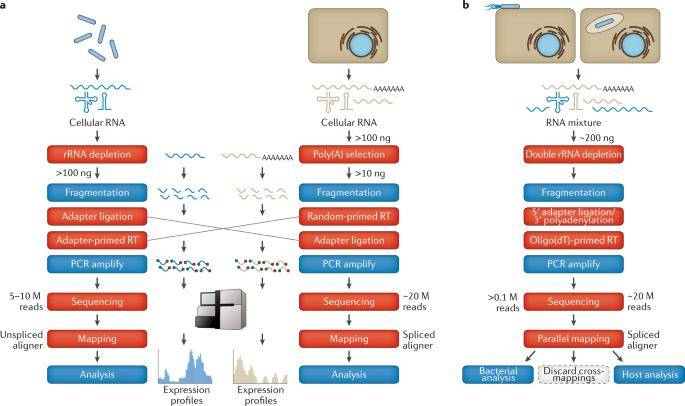 The basic steps in commonly used protocols for strand-specific bacterial, mammalian or dual expression profiling. (Westermann et al., 2021)
The basic steps in commonly used protocols for strand-specific bacterial, mammalian or dual expression profiling. (Westermann et al., 2021)
Single-Cell Transcriptome Sequencing (scRNA-seq)
In recent years, the advancement of single-cell transcriptome sequencing (scRNA-seq) technology has ushered in a new era of transcriptomics, leading to numerous significant discoveries. These discoveries range from the identification of previously unknown cell types and physiological states to the unraveling of novel gene regulatory pathways. Researchers have begun to recognize the pivotal role of cellular heterogeneity in host-pathogen interactions. For instance, pathogens exploit pre-existing and pre-induced cellular heterogeneity to establish infectious ecological niches.
Within the pathogen population itself, there exists substantial phenotypic diversity. For example, Salmonella typhimurium exhibits a bistable expression of invasive genes even before encountering host cells, and this gene expression diversity intensifies throughout the course of infection. This strategy offers a unique advantage - the invasion of certain subpopulations of Salmonella induces epithelial inflammation, which in turn facilitates the further invasion of other Salmonella typhimurium strains in the intestinal lumen.
To gain deeper insights into these intricate dynamics, higher-resolution transcriptomic approaches are imperative. These advanced methods enable us to capture and dissect how cellular heterogeneity influences the outcomes of host-microbe interactions.
Reference:
- Westermann, Alexander J., and Jörg Vogel. "Cross-species RNA-seq for deciphering host–microbe interactions." Nature Reviews Genetics 22.6 (2021): 361-378.


 Sample Submission Guidelines
Sample Submission Guidelines
