Strategies to Mitigate Common DNA Contamination in Microbial Sequencing
The field of microbial community research has transformed our comprehension of microbiology, yet the pervasive issue of DNA contamination can compromise the integrity of experiments, from initial sampling to the final sequencing stage. Contaminants may infiltrate DNA samples during various steps of microbial research, including DNA extraction, PCR amplification, and even laboratory reagents, ultimately distorting the results. In particular, DNA contamination can be detrimental when studying samples with minimal microbial loads, necessitating careful measures to address this challenge.
Sources of DNA Contamination
The potential sources of DNA contamination are manifold and encompass molecular biology-grade water, PCR reagents, and the DNA extraction kits themselves. To discern and quantify contamination levels, negative controls, often referred to as "blank" DNA extraction and subsequent PCR amplification samples, are routinely included in microbial sequencing experiments. Interestingly, these negative control samples typically reveal a spectrum of contaminating bacterial species, mirroring the background found in samples of human origin processed alongside the same batch of DNA extraction kits.
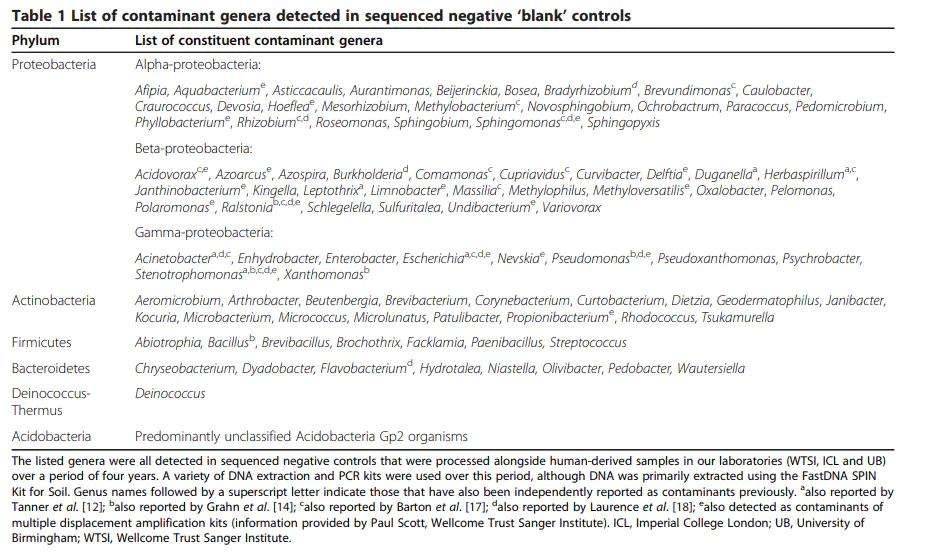
An examination of these negative control results frequently uncovers common taxonomic classifications across different laboratory locations. Among the recurring contaminants are Acidobacteria Gp2, Burkholderia, Mesorhizobium, and Pseudomonas. These similarities suggest that variations in contaminant levels exist between laboratories, potentially arising from differences in reagents, kit batches, or even contaminants introduced within the laboratory environment.
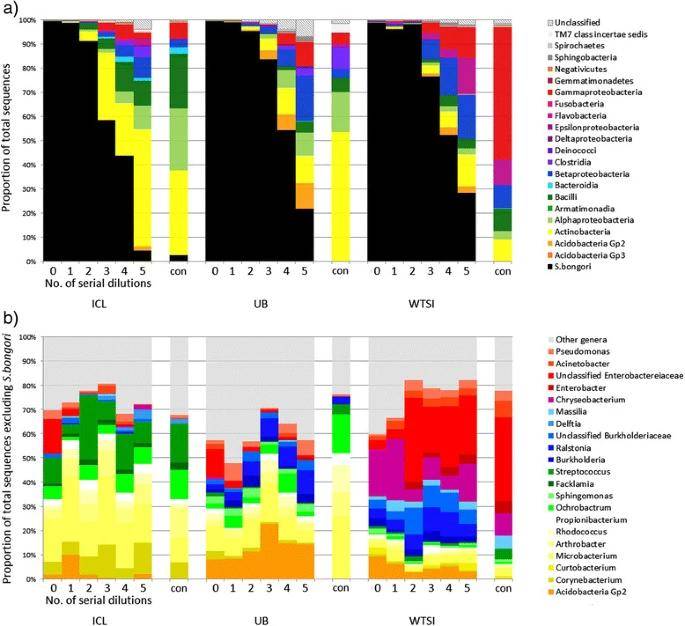 Summary of 16S rRNA gene sequencing taxonomic assignment from ten-fold diluted pure cultures and controls. (Salter et al., 2014)
Summary of 16S rRNA gene sequencing taxonomic assignment from ten-fold diluted pure cultures and controls. (Salter et al., 2014)
Impact of Contaminated Extraction Kits on Low Biomass Studies
To illustrate the severity of the issue, let's consider a specific case. Nasopharyngeal swabs were employed to investigate the evolution of nasopharyngeal microbiota in infants. Principal Coordinate Analysis (PCoA) unveiled two distinct taxa, with samples collected during the first six months of life clearly distinguishable from those taken at later stages. This finding underscores the correlation between nasopharyngeal bacterial flora and developmental time. However, a crucial variable emerged when four different batches of DNA extraction kits were employed. The choice of kit markedly influenced the community characteristics of the samples, with the majority of sample reads originating from the first two kits used.
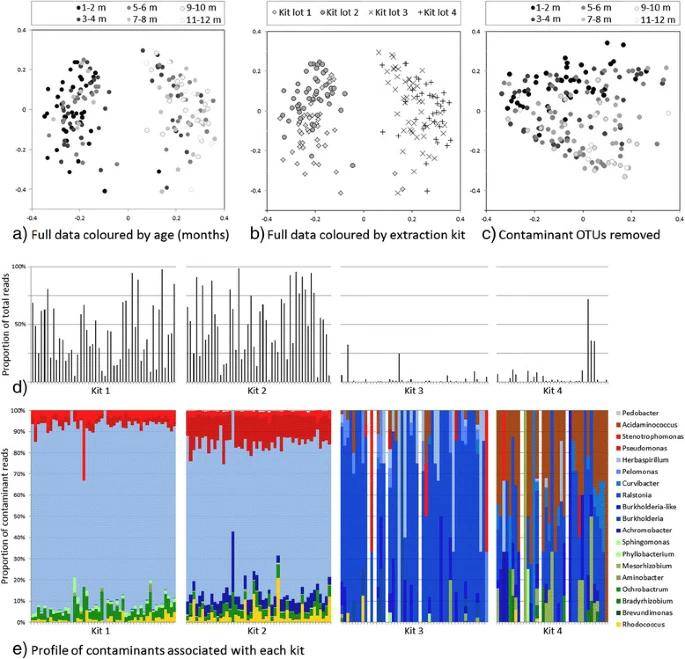 Summary of the contaminant content of nasopharyngeal samples from Thailand. (Salter et al., 2014)
Summary of the contaminant content of nasopharyngeal samples from Thailand. (Salter et al., 2014)
Recommendations
Given the profound implications of bacterial DNA contamination on microbial studies, especially when dealing with samples of low microbial biomass, a set of key recommendations emerges:
Kit Consistency: To minimize contamination introduced by the kits themselves, it is strongly advised to employ the same batch of DNA extraction kits consistently throughout a project. This practice ensures uniformity and eliminates the risk of kit-induced variation in results.
Conclusion
The battle against DNA contamination in microbial sequencing is an ongoing endeavor, vital for the reliability and reproducibility of microbiota studies. By understanding the sources of contamination, recognizing its impact on results, and implementing measures like kit consistency, researchers can safeguard the integrity of their microbial community research, even when dealing with samples of low microbial biomass. This approach will ultimately lead to more accurate and dependable findings, further advancing our understanding of microbiology.
Reference:
- Salter, Susannah J., et al. "Reagent and laboratory contamination can critically impact sequence-based microbiome analyses." BMC biology 12 (2014): 1-12.


 Sample Submission Guidelines
Sample Submission Guidelines
