A Comprehensive Exploration of Methods for Analyzing Integration Sites
Gene and Cell Therapy
In 1990, W.F. Anderson, a pioneering American medical scientist, achieved a groundbreaking feat in the realm of gene therapy. His transformative work involved the application of gene therapy to a 4-year-old girl grappling with adenosine deaminase deficiency, marking a historic moment as the world's inaugural triumph in gene therapy and a pivotal milestone in medical history. In the subsequent decades of relentless development, gene therapy has emerged as a beacon of hope, offering a paradigm shift in the approach to treating ailments by addressing them at their genetic roots.
Categorically, gene therapy unfolds in two distinct modes of treatment. In vivo therapy entails the direct injection of vectors bearing the required genes into the bloodstream or specific target organs. This method finds application in the realms of monogenic genetic diseases, hemophilia, and ophthalmic conditions. On the other hand, in vitro therapy, recognized as cell therapy, involves the extraction of a patient's cells, genetic modification outside the body, and subsequent reintroduction into the patient's system. One noteworthy manifestation of in vitro therapy is Chimeric Antigen Receptor T-cell therapy (CAR-T), a cutting-edge precision-targeted approach heralding a new era in tumor treatment.
The trajectory of progress is evident in the approval of over 20 gene therapy drugs and 8 CAR-T products by national drug regulatory agencies. Armed with sophisticated biotechnology and promising therapeutic avenues, gene and cell therapy medications are poised to play a pivotal role in the future landscape of disease prevention and treatment.
CD Genomics' lentiviral integration sites analysis service utilizes the power of Illumina Platform for integration sites detection, and has developed a low-cost high-throughput approach by using target enrichment sequencing or LM-PCR method. We are pleased to use our extensive experience and advanced platform to offer the best service and the most qualified products to satisfy each demand from our customers.
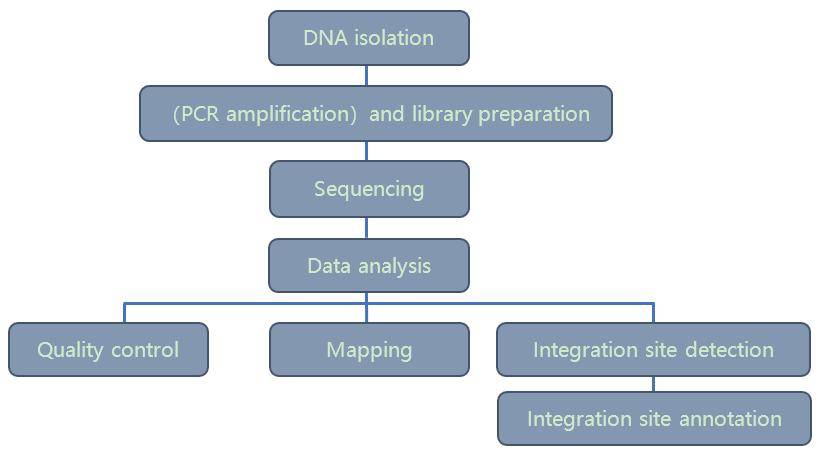 Integration site analysis workflow – CD Genomics
Integration site analysis workflow – CD Genomics
Lentiviral Integration in Gene and Cell Therapy
In the realm of gene and cell therapy, the mode and efficacy of gene delivery play a pivotal role in determining the therapeutic impact of the drug. Consequently, the selection of an appropriate vector assumes paramount importance. Lentiviral systems have garnered widespread use in diverse therapeutic applications owing to their expansive infectivity spectrum, adept infection of both dividing and non-dividing cells, and the sustained and stable expression of exogenous genes over the long term.
Viral integration stands as a crucial facet of the standard lentiviral life cycle. The process commences with the virus recognizing a receptor on the host cell's surface, facilitating the transfer of RNA and proteins into the cell. Following this, RNA undergoes reverse transcription to DNA, and the integrase recognizes the Long Terminal Repeat (LTR) carried by the viral DNA. It then cleaves the host genome in a random or semi-random fashion, resulting in sticky ends that seamlessly fuse with the viral DNA, completing the insertion of the exogenous gene. Viral integration emerges as an indispensable step for the expression of exogenous genes and the actualization of the therapeutic effects of gene therapy.
Methods for Analyzing Integration Sites
- PCR Technology
Originally, the analysis of viral integrations involved Southern blot and genomic libraries to characterize integration sites. Over time, PCR has supplanted Southern blot as the primary analytical method for viral integrations due to its operational simplicity and result accuracy. Within the realm of PCR technology, three main categories have emerged for viral integration analysis: Reverse PCR (R-PCR), Ligated Mediated PCR (LM-PCR), and Linear Amplification Mediated PCR (LAM-PCR).
In Reverse PCR, DNA is enzymatically cleaved and then ligated into a loop. Reverse primers are designed for amplification, allowing for the retrieval of unknown sequences on both sides of the viral integration site. However, the limited efficiency of ligase loop formation constrains the utility of reverse PCR.
LM-PCR involves connecting the two ends of DNA after cleavage and interruption, with primers designed based on fixed sequences of viral DNA and joints. This approach unlocks the positional information of the viral integration site. LM-PCR is known for its simplicity and ease of operation, remaining in use from its inception to the present day.
LAM-PCR represents a more advanced technique where DNA is initially linearly amplified, and the single-stranded product obtained from this amplification is further amplified by LM-PCR to yield the specific product of the single-stranded amplification. The introduction of linear amplification enhances the sensitivity and specificity of LAM-PCR, surpassing that of Reverse PCR and LM-PCR, making it a widely adopted method in current research.
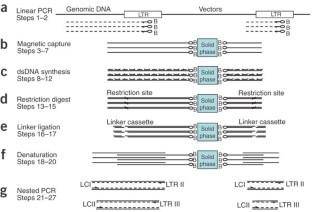 Schematic outline of LAM-PCR to amplify 5'-LTR retroviral vector-genomic fusion sequences. (Schmidt et al., 2007)
Schematic outline of LAM-PCR to amplify 5'-LTR retroviral vector-genomic fusion sequences. (Schmidt et al., 2007)
- Next-Generation Sequencing (NGS)
Traditional PCR technology, reliant on restriction endonuclease digestion for integrative site analysis, has inherent preferences. Simultaneously, electrophoresis or first-generation sequencing, employed in the past, suffers from low sensitivity and a limited capacity for analyzing clones. The landscape shifted in 2005 with the introduction of the first high-throughput sequencer, Roche 454, marking the ascent of next-generation sequencing (NGS) as the predominant technology for integrative site analysis. NGS, owing to its copious sequencing data, significantly broadens the spectrum of integration sites. Notably, it excels in detecting low-frequency integration clones, furnishing invaluable insights for drug development.
The process of constructing high-throughput sequencing libraries is more intricate compared to traditional PCR. Commonly used library construction methods include LM-PCR, LAM-PCR, and non-restriction enzyme digestion LAM-PCR. While LAM-PCR library construction involves the complexities and high costs associated with biotin-labeled primers and streptavidin-labeled magnetic beads in linear amplification, LM-PCR emerges as a pragmatic alternative. LM-PCR facilitates the enrichment of insertion site sequences through a two-step PCR process, enabling library construction in 6-7 hours. This streamlined approach accelerates the library construction cycle, making it an optimal method for detecting lentiviral insertion sites.
Lentiviruses possess long terminal sequences (5'LTR and 3'LTR) at each end, and LM-PCR can amplify both LTRs to form libraries. However, in conventional library construction methods, half of the downstream data reads become redundant information concerning the integration of LTRs with proviral sequences. Thus, adapting LM-PCR stands out as an efficient and ideal choice for lentivirus insertion site detection.
Reference:
- Schmidt, M., Schwarzwaelder, K., Bartholomae, C. et al. High-resolution insertion-site analysis by linear amplification–mediated PCR (LAM-PCR). Nat Methods 4, 1051–1057 (2007).


 Sample Submission Guidelines
Sample Submission Guidelines
