oxBS-Seq, An Epigenetic Sequencing Method for Distinguishing 5mC and 5mhC
In order to distinguish between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), oxidative bisulfite sequencing (oxBS-Seq) provides an extra oxidative stage to bisulfite. Initially, 5hmC is oxidized in genomic DNA to 5-formylcytosine (5fC). Unlike 5mC and 5hmC, 5fC is delicate to bisulfite deamination, so the oxidized DNA bisulfite treatment transforms 5fC to uracil. The cytosines remaining in the sequence were 5mC and not 5hmC utilizing next sequencing. Parallel identification of 5mC and 5hmC is conducted in a standard bisulfite run. The involvement of 5hmC may be deduced by contrasting these two sequences. oxBS-Seq's main advantage over enzymatic methods such as TAB-seq is that oxBS-Seq does not involve a heavily engaged TET enzyme that can be costly and only about 95% effective.
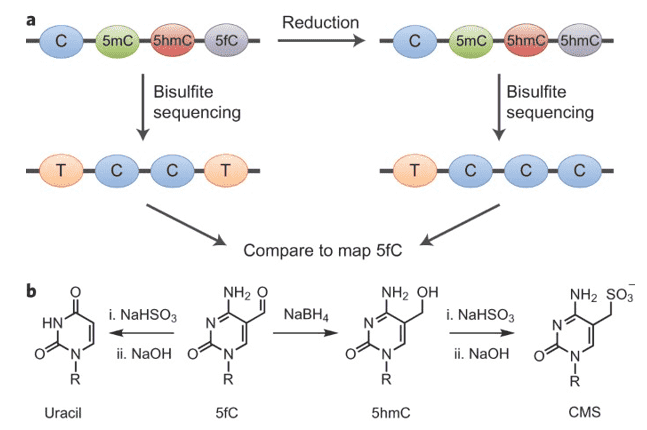
Figure 1. Quantitative sequencing of 5-formylcytosinein DNA at single-base resolution. (Booth, 2014)
Advantages and Disadvantages of Oxidative bisulfite Sequencing
There are three main benefits to oxBS-Seq. These involve (1) CpG and non-CpG methylation at single-base resolution throughout the genome, (2) covering 5mC in dense and less thick repeat areas, and (3) clearly distinguish between 5mC and 5hmC.
But oxBS-Seq has its disadvantages as well. Its two existing disadvantages are (1) significant DNA loss (99.5 percent) that can result from severe oxidation circumstances, and (2) it must be integrated with bisulfite sequencing (BS-Seq) to differentiate and evaluate C, 5mC, and 5hmC.
Principles of oxBS-Seq
5mC is an epigenetic DNA symbol that is enhanced with CpG dinucleotides and performs an essential part in gene silencing and genome reliability. 5mC is a popular epigenetic mark with significant mechanisms in growth, transposon and gene silencing, genomic imprinting, inhibition of X-chromosomes, and sustainability of the genome. In many human illnesses, such as cancer, in which gross hypomethylation is followed by hypermethylation within certain CpG islands, abnormal DNA methylation has been involved.
The ten-eleven translocation (TET) enzyme family can oxidize 5mC into 5hmC. In active DNA demethylation, 5hmC may be an alternative but could also involve an epigenetic mark per se. By analytical techniques, rates of 5hmC in genomic DNA can be computed and plotted by enhancement of 5hmC-containing DNA portions that are then sequenced. Such methods have comparatively low resolution and provide only relative quantifiable information. Using BS-Seq, single nucleotide sequencing of 5mC was carried out, but 5mC from 5hmC cannot be discriminated against by this technique. Derivatized 5hmC in genomic DNA can be detected by single-molecule real-time sequencing (SMRT). However, the enhancement of DNA fragments involving 5hmC is necessary, causing the failure of quantitative data. In addition, SMRT has a significantly high rate of sequencing errors, and modifying peak calling is inaccurate. 5mC of 5hmC can be resolved by protein and solid-state nanopores and can sequence unamplified DNA.
Via extremely selective chemical oxidation of 5hmC to 5fC, oxBS-Seq distinguishes between 5mC and 5hmC. The method also utilizes the bisulfite distinction in reactivity between 5hmC and 5fC. Bisulfite treatment creates 5fC to be deformylated and deaminated to establish uracil (which is read as thymine at the sequencing stage) as opposed to 5hmC which does not deaminate. Thus, after oxBS-seq, the only base that is not deaminated and therefore read as a cytosine is 5mC. Correspondingly, this strategy provides a reliable overview of all the current 5mC. By conducting BS-seq and evaluating its findings with those of oxBS-seq, subsequent identification of 5hmC is attained.
Applications and Research Progress
Together with a multitude of analytical techniques that have already been established for BS-seq, such as DNA sequencing and methylation arrays, the oxBS-seq process can be employed to alter DNA. This technique is system-agnostic when utilized in conjunction with next-generation sequencing and is suitable for the assessment of 5mC and 5hmC at single-base resolution in both whole-genome or targeted-region platforms. oxBS-seq is suitable with enhancement and targeting methods, such as post-bisulfite locus-specific PCR and BS-seq lowered representation, in addition to whole-genome evaluation.
References:
- Kirschner K, Krueger F, Green AR, Chandra T. Multiplexing for Oxidative Bisulfite Sequencing (oxBS-seq). InDNA Methylation Protocols 2018. Humana Press.
- Ecsedi S, Rodríguez-Aguilera JR, Hernandez-Vargas H. 5-Hydroxymethylcytosine (5hmC), or how to identify your favorite cell. Epigenomes. 2018, 2(1).
- Booth MJ, Marsico G, Bachman M, et al. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nature chemistry. 2014, 6(5):435.
- Booth MJ, Branco MR, Ficz G, et al. Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012.
- Schüler P, Miller AK. Sequencing the Sixth Base (5‐Hydroxymethylcytosine): Selective DNA Oxidation Enables Base‐Pair Resolution. Angewandte Chemie International Edition. 2012, 51(43).


 Sample Submission Guidelines
Sample Submission Guidelines
