Multi-omics Analysis Reveals The Pathogenesis of Cancer
Multiomics focuses more on the collection of complete cell information and the attention to time and space, so it can better and more comprehensively reflect the characteristics of cells compared with single cell sequencing technology. In addition, it also can provide more accurate biological insights and a more systematic understanding of biological heterogeneity and diversity. In conclusion, the multi-group data provide researchers with different perspectives, deepening the understanding of different biological pathways in the process of tumor progression and providing new insights for exploring the mechanism of tumor regulation and determining potential therapeutic targets.
Single-cell multi-omics analysis reveals regulatory programs in clear cell renal cell carcinoma
In recent years, a number of single-cell multi-group technology based on the development of single-group technology have emerged. Single-cell multi-omics technology can capture and analyze multiple omics information in the same cell, including single-cell transcriptome, genome, epigenome, proteome and other aspects of information. Clear cell renal carcinoma (ccRCC) is the most common and aggressive histological subtype of renal cell carcinoma. Due to its highly complex tumor microenvironment heterogeneity, the existing clinical intervention strategies, such as targeted therapy and immunotherapy have not achieved good therapeutic effects. So, it is urgent to understand the underlying cellular and molecular mechanisms to facilitate the discovery of biomarkers and guide clinical intervention. Researchers applied single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) to generate transcriptional and epigenomic landscapes of ccRCC. They identified tumor cell-specific regulatory programs mediated by four key transcription factors (TFs), which have prognostic significance in The Cancer Genome Atlas (TCGA) database. Taken together, these multiomics approaches further clarify the cellular heterogeneity of ccRCC and identify potential therapeutic targets. Meanwhile, multiomics joint analysis delineated the intercellular communications mediated by ligand–receptor interactions within the tumor microenvironment.
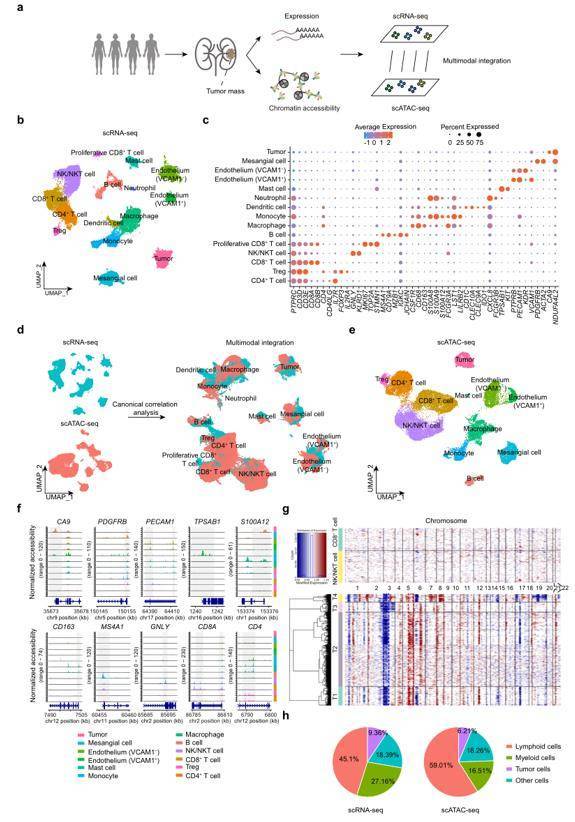 Single-cell transcriptional and chromatin accessibility profiling of human ccRCC
Single-cell transcriptional and chromatin accessibility profiling of human ccRCC
Transcriptomics, proteomics, and lipidomics analysis reveal the mechanism of tumor inhibition
Oral and esophageal squamous cell carcinomas (SCCs) are associated with high mortality, yet the molecular mechanisms underlying these malignancies are largely unclear. Recent studies have revealed that OTUB2/STAT1/CALML3/PS axis has anti-tumor effect through transcriptomics, proteomics, lipidomics and molecular biology experiments. Multigroup integration analysis showed that this function was realized by promoting the phosphorylation and dimerization of transcription factor STAT1, and then activating CALML3 transcription. Phosphatidylserine synthesis is a key metabolic event that mediates OTUB2's anti-tumor function, and preclinical research results suggest that oral phosphatidylserine may help to inhibit the occurrence of upper gastrointestinal squamous cell carcinoma. This study shows the potential of PS administration as a strategy for the treatment and prevention of tongue and esophageal SCCs.
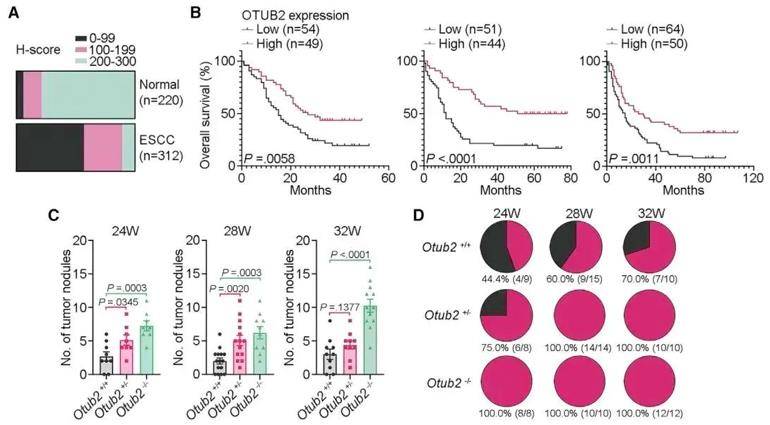 OTUB2 showed tumor inhibition
OTUB2 showed tumor inhibition
Whole-exome and targeted sequencing identify potential drivers of tumor recurrence
Breast cancer and ovarian cancer have a high incidence rate among women. They are malignant tumors with long latency, no obvious early onset symptoms, and high mortality. Pathogenic variants in BRCA1/2 are the most common cause of hereditary breast and ovarian cancer. The loss of BRCA1 or BRCA2 allows cells to accrue mutations and genomic rearrangements, which facilitate a transition to cancer. Researchers Identify potential recurrence-specific drivers by performing multi-omic sequencing on 67 paired primary and recurrent BrCa and OvCa from 27 BRCA1/2 mutation carriers. The systematic analysis finds two BRCA2 isoforms, BRCA2-201/Long and BRCA2-001/Short, BRCA2-001/Short is expressed more frequently in recurrences and related to reduced overall survival in breast cancer. This study revealed multiple potential drivers of recurrent diseases in BRCA1/2 mutation related cancers, improving our understanding of tumor evolution and proposed potential biomarkers.
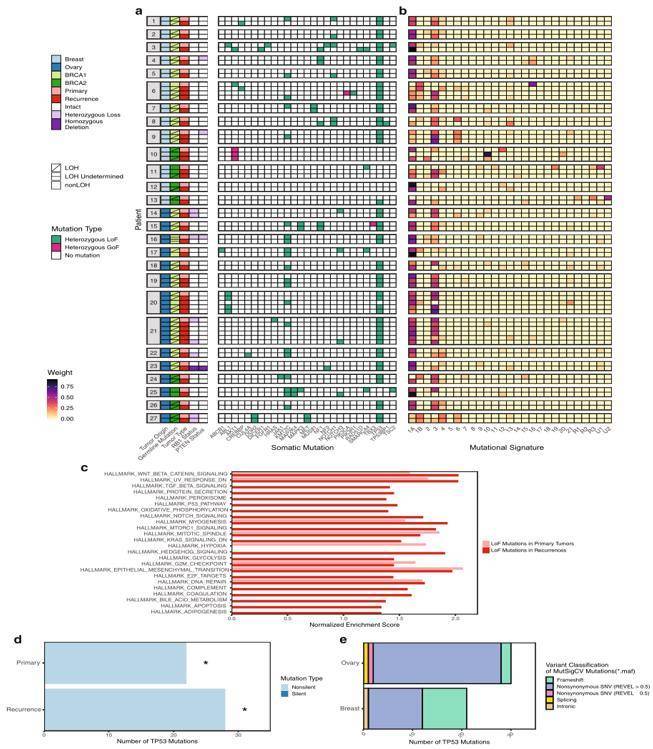 Integrated analysis of somatic mutations by whole-exome and targeted sequencing
Integrated analysis of somatic mutations by whole-exome and targeted sequencing
At present, multi-omics technology is booming and playing an important role in the field of tumor research. The data set generated by these technologies will help to better understand tumor genesis, tumor progression, tumor microenvironment changes, and key biological mechanisms of drug resistance. With the further development of multi-omics technology in the future, we firmly believe that it will be more widely used in tumor research and help the development of tumor precision medicine.
References:
- Shah JB, Pueschl D, Wubbenhorst B, et al. Analysis of matched primary and recurrent BRCA1/2 mutation-associated tumors identifies recurrence-specific drivers. Nat Commun. 2022;13(1):6728. Published 2022 Nov 7.
- Zhou Y, Bian S, Zhou X, et al. Single-Cell Multiomics Sequencing Reveals Prevalent Genomic Alterations in Tumor Stromal Cells of Human Colorectal Cancer. Cancer Cell. 2020;38(6):818-828.e5.
- Chang W, Luo Q, Wu X, et al. OTUB2 exerts tumor-suppressive roles via STAT1-mediated CALML3 activation and increased phosphatidylserine synthesis. Cell Rep. 2022;41(4):111561.
- Long Z, Sun C, Tang M, et al. Single-cell multiomics analysis reveals regulatory programs in clear cell renal cell carcinoma. Cell Discov. 2022;8(1):68. Published 2022 Jul 19.


 Sample Submission Guidelines
Sample Submission Guidelines
