Since bacteria lack histones and nucleosomes, they primarily use DNA methylation as their epigenetic gene regulation. There are three different types of DNA methylation in bacterial genomes: N4-methylcytosine (4mC), N6-methyladenine (6mA), and 5-methylcytosine (5mC). Generally, prokaryotes have higher 6mA levels but lower 5mC levels than plants and mammals. DNA is methylated when a methyl group from S-Adenosyl-L-Methionine (SAMe) is transferred by the methyltransferase (MTase) enzyme on target bases.
The distribution of 6mA in the whole genome varies between eukaryotes and prokaryotes. N6-methyladenine is lesser observed and lesser motif-driven in eukaryotes than prokaryotes. This reduced number of 6mA methylation in eukaryotes could be due to evasion of modified 6mA sites from cognate restriction enzymes practically hiding its presence at specific sequence motifs. Furthermore, unlike eukaryotes, prokaryotic DNA is not packaged in nucleosomes thus exposing a larger number of motifs to be methylated. Despite 6mA differences between eukaryotes and bacteria, the intrinsic properties and their effect on DNA conformation are observed to be consistent.
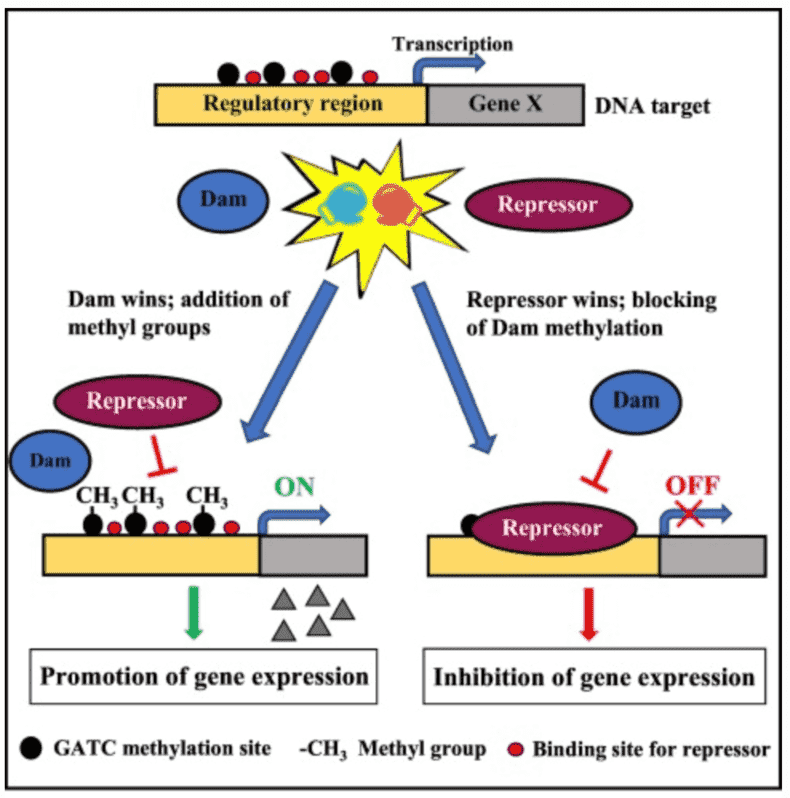
Some biological functions of 6mA methylation include alteration of DNA double helix formation through base stacking and base-pair stability; when 6mA occurs after a T, DNA is observed to be overwound and bent; discriminate self from foreign DNA in restriction-modification systems; distinguish the parental DNA strand from the daughter strand with detected mutations, and marks regions for DNA replication initiation and alters the rate of cell cycle progression.
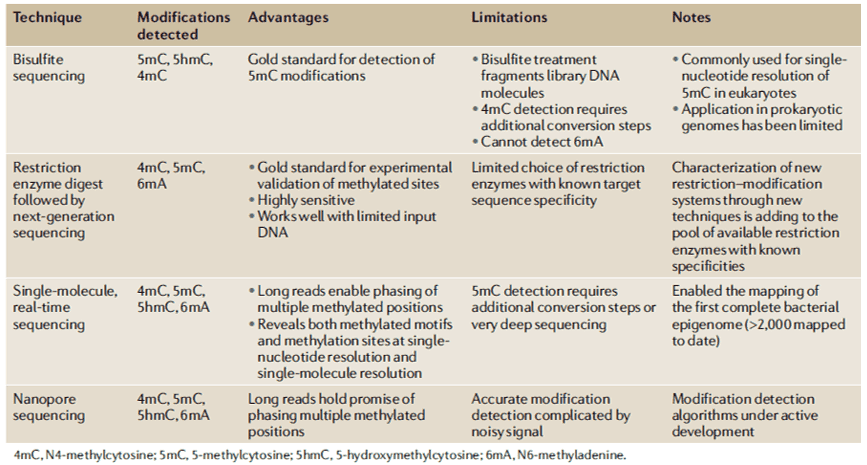
Several methods have been created to detect 6mA levels. In determining the specific genomic locations of 6mA, MeDIP with microarray analysis (MeDIP-seq) was developed to detect near single-nucleotide 6mA resolution; however, it is antibody-dependent. Furthermore, single-molecule real-time sequencing (SMRT-seq) is also being used for loci detection. SMRT-seq is a third-generation sequencing technique that measures the kinetic rate of nucleotide incorporation during sequencing making the identification of every DNA modification at single-base resolution. When coupled with UHPLC-MS/MS, they can give an accurate confirmation of both the genomic location and presence of 6mA in an organism.
In the recent studies involving genome-wide mapping study of 6mA in bacteria, traditional methods such as RM digestion-based approaches were used. However, the majority of 6mA methylation studies have already adopted SMRT sequencing. Through the SMRT sequencing, the vast diversity of 6mA MTases and methylated motifs in bacteria were elucidated including the unknown MTases and previously uncharacterized RM systems. Upon functional knockout of certain 6mA MTases, most of the organisms exhibited widespread changes in transcriptional processes. On the other hand, other studies highlight that differently methylated 6mA positions influences by varying growth stages and environment. Bacteriophage invasion and host resistance have been also studied through SMRT sequencing. In a study, 936-type bacteriophages were shown to encode various 6mA MTases which renders the bacteriophages resistant with protective methylation against the host RM systems. For bacterial DNA methylation mapping, SMRT sequencing, and the Oxford Nanopore sequencing holds huge potential for methodological advancement.
References
- Beaulaurier J, Schadt EE, Fang G. Deciphering bacterial epigenomes using modern sequencing technologies. Nature Reviews Genetics. 2019, 20(3).
- O'Brown ZK, Greer EL. N6-methyladenine: a conserved and dynamic DNA mark. InDNA Methyltransferases-Role and Function, 2016 (pp. 213-246). Springer, Cham.