Introduction to Microbial Epigenetics
Epigenetics has been defined as the study of stable alterations in gene expression potentials that arise during development and cell proliferation, or alterations in DNA function without alterations in DNA sequence. Modern epigenetic features refer to the alteration of DNA and/or associated proteins without nucleotide sequence variance, which transmits the data contained to the next generation. It has been considered that diseases are generally influenced by obtained biological alterations, but it is becoming evident that any phenotype is the result of a complex interplay between genotype, epigenome, and environment. Epigenetics concentrates on pathways that control how and when specific genes are turned on and/or off, while epi-genomics assess epigenetic changes in a cell or whole organism across many genes.
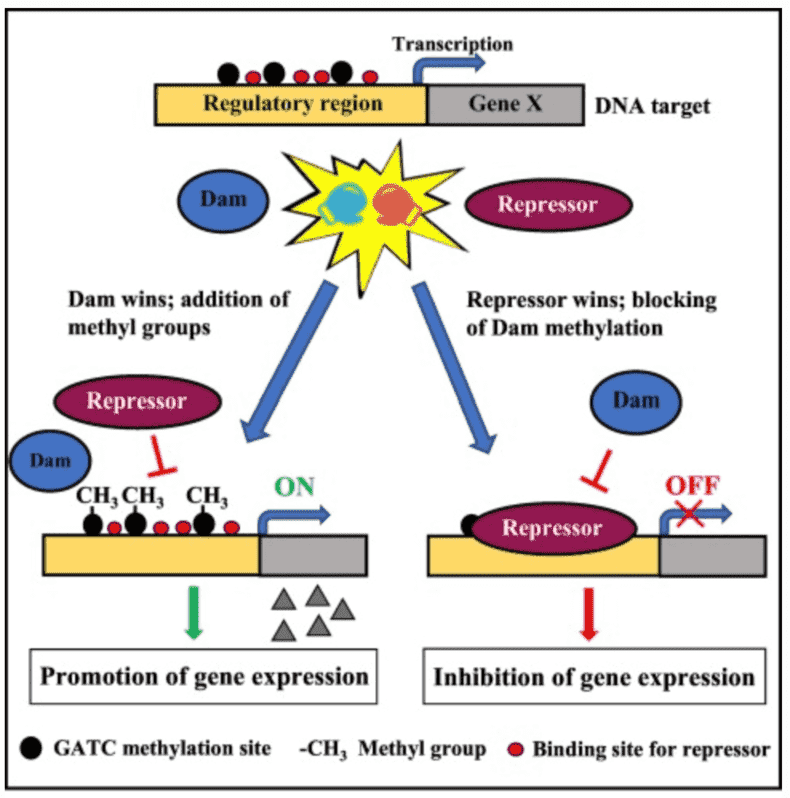 Figure 1. DNA methylation-dependent gene expression regulation in bacteria
Figure 1. DNA methylation-dependent gene expression regulation in bacteria
From one cell division to the next, epigenetic DNA and chromatin modifications prevail and can happen for several generations of cells. Biomarkers for the detection of certain metabolic diseases could be methylated DNA, acetylated proteins, miRNA, specific substrates, cofactors, enzymes engaging in biochemical reactions linked to epigenomic procedures in body fluids.
Epigenetic alterations induced by microbial invasion
Numerous studies have exhibited that specific pathogens (Helicobacter pylori, Streptococcus bovis, Chlamydia pneumoniae, Campylobacter rectus, Epstein-Barr virus, hepatitis virus, human papillomavirus, polyomavirus, etc.) may result in the epigenetic variations of the host, leading to the onset and progression of certain diseases, particularly malignancies. The mutualistic microorganisms of coral reef cnidarian symbiosis, pea aphids, and cactuses have been involved in epigenetic thermotolerance differences.
As obligate intracellular parasites, microbial manipulation of host epigenetic marks has helped to enhance numerous ways of hijacking the cell system to ensure the implementation of their life cycle and sometimes to escape their host's immune responses. Microbes that cause prolonged infections are likely to gain from inherited host transcription epigenetic changes that create an environment for their latent or continuous state without the inciting effectors having to be constantly expressed. As it aims for such epigenetic control, host genes involved in cell cycle progression, senescence, survival, inflammation, and immunity are the main targets. Some severe bacterial infections are also linked with malignancy, with the human gastric mucosa disease of Helicobacter pylori being the most thoroughly researched. In addition, many microbes have progressed ways of avoiding the immune response, and these mechanisms have again involved gene mutations in host cells.
DNA methylation is being used by most epigenetic processes known in bacteria as a sensor that controls a particular connection between DNA and protein. Generally, these systems consist of DNA methylase and DNA binding protein(s) that connect to DNA sequences that coincide with the target methylation site, obstructing that site's methylation. In turn, methylation of the target site restricts protein binding, leading in methylated and non-methylated, two optional methylation states of the target site. Mechanisms unassociated with the transmission of bacterial restriction-modification systems appear to instigate the epigenetic modifications caused by contaminating bacteria in multicellular eukaryotes and portray a novel research field to be researched.
Pathogen-induced modifications of the host
In the scientific literature, pathogen-induced modifications in host physiology, morphology, and behavior are thoroughly reported. Perhaps the most fascinating examples of these modifications are those that have been shown to be the outcome of the pathogen's deceptive tactic to maximize its survival and transmission. Even so, evidence has emerged in the last few years that histone modifications and chromatin remodeling enforce gene expression and are therefore key targets during infection for pathogen manipulation. The immune system of the host is one such evident target. In recent years, as a comparably regular sight of pathogenic viral and bacterial infections, epigenetic modification of the host transcriptional program connected to host defense genes has surfaced. Bacteria are the cornerstone of microbe epigenetic studies and supply several pioneering instances of host gene reprogramming caused by contamination.
References
- Riber L, Hansen LH. Epigenetic Memories: The Hidden Drivers of Bacterial Persistence?. Trends in Microbiology. 2021 Jan 4.
- Sánchez-Romero MA, Casadesús J. The bacterial epigenome. Nature Reviews Microbiology. 2020 Jan;18(1).
- Al Akeel R. Role of epigenetic reprogramming of host genes in bacterial pathogenesis. Saudi journal of biological sciences. 2013 Oct 1;20(4).