Metagenomic Next-generation Sequencing Promotes The Detection of Respiratory Pathogens
Recently, metagenomic next-generation sequencing (mNGS) was developed and shows its superiority in terms of disease diagnosis. Infection is one of the main causes of death in critically ill patients.With the emergence of new and drug-resistant pathogens and the gradual increase of immunosuppressive population, the incidence rate and mortality of infection remain high. Among them, respiratory tract infection is one of the most common types of infection. mNGS has been widely used in clinic for its wide coverage and large amount of information in detecting pathogens of respiratory tract infection.
Application of mNGS in Pathogenic Diagnosis of Respiratory Tract Samples
The application of clinical mNGS for diagnosing respiratory infections improves etiology diagnosis. Researchers perform strategy evaluation and metagenomic analysis for the mNGS data generated between March 2017 and October 2019. Concurrently, the diagnostic intensity of four sample types was evaluated to determine the type more suitable for the diagnosis of respiratory tract infection by mNGS. The result showed that the positive predictive values (PPVs) for mNGS diagnosing of non-mycobacterium, Nontuberculous Mycobacteria (NTM), and Aspergillus were obviously higher in bronchoalveolar lavage fluid (BALF) demonstrating the potency of BALF in mNGS diagnosis. Lung tissue and sputum can be selected as samples when suspected of Mycobacterium tuberculosis infection; The immune status of the host is also one of the factors that affect the structure of respiratory tract microorganisms, and the detection rate of herpes virus in immunocompromised hosts is higher. This bioinformatic pipeline successful helps mNGS data interpretation and reduces manual corrections for etiology diagnosis.
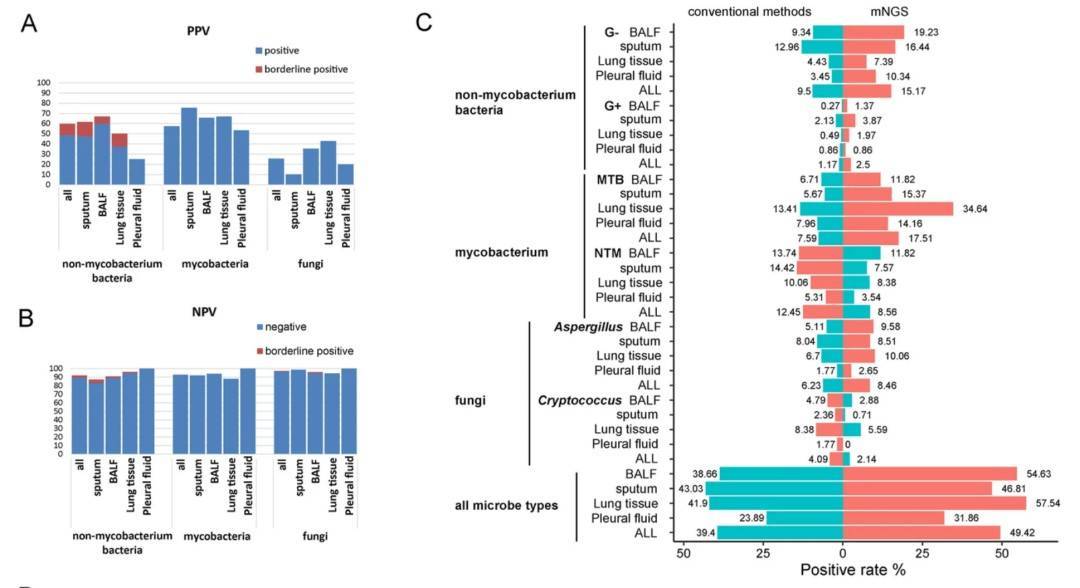 Evaluation of the mNGS performance in four respiratory specimen types and multiple pathogen categories.
Evaluation of the mNGS performance in four respiratory specimen types and multiple pathogen categories.
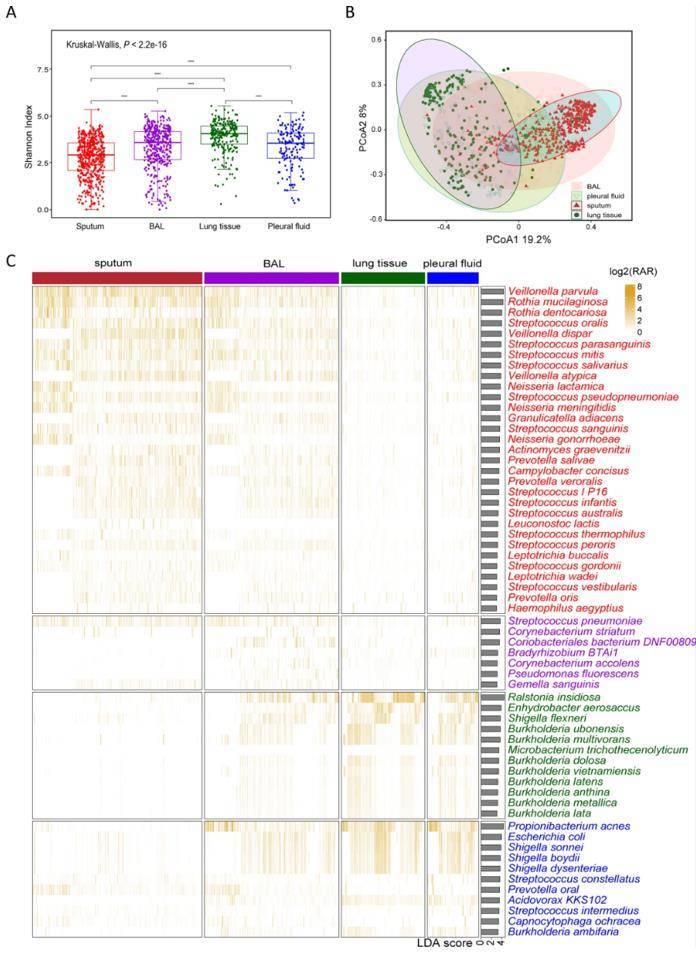 Distribution of respiratory tract flora of mNGS in different sample types
Distribution of respiratory tract flora of mNGS in different sample types
Clinical application of macrogenomic sequencing in the diagnosis of lower respiratory tract infection
Lower respiratory tract infections (LRTIs) are the most common cause of death in low-income countries. Macrogenome second generation sequencing (mNGS) has high throughput and can detect multiple pathogens in respiratory tract samples at the same time, which makes up for the limitations of traditional microbial detection methods and has been applied to the detection of clinical samples. The study collected bronchoalveolar lavage fluid samples from 162 patients, and evaluated the diagnostic efficacy of mNGS in LRTIs by combining microbiological detection and comprehensive reference standards. At the same time, transcript analysis was carried out on the host of infected/uninfected group, bacterial/viral group and tuberculosis/non-tuberculosis group to explore the markers of LRTIs, viral LRTIs and tuberculosis. The results showed that the sensitivity of mNGS detection could be improved by optimizing DNA extraction and enriching sample DNA concentration. The five DEGs involved in lower respiratory tract infection found in this study are related to the immune response to infection. However, due to the relatively small sample size and the preliminary nature of this analysis, whether these genes can be used as markers to predict LRTIs needs further research and verification. It is important that his study confirmed the advantages of mNGS in the detection and identification of LRTIs pathogenic bacteria. MNGS can detect pathogens that are difficult to cultivate, which makes up for the shortcomings of traditional culture methods. Compared with traditional detection methods, mNGS can detect more pathogenic microorganisms, and the clinical application of mNGS will help the antimicrobial management of LRTIs. At the same time, mNGS broadens the pathogen detection spectrum of LRTIs, which is helpful for targeted treatment of patients.
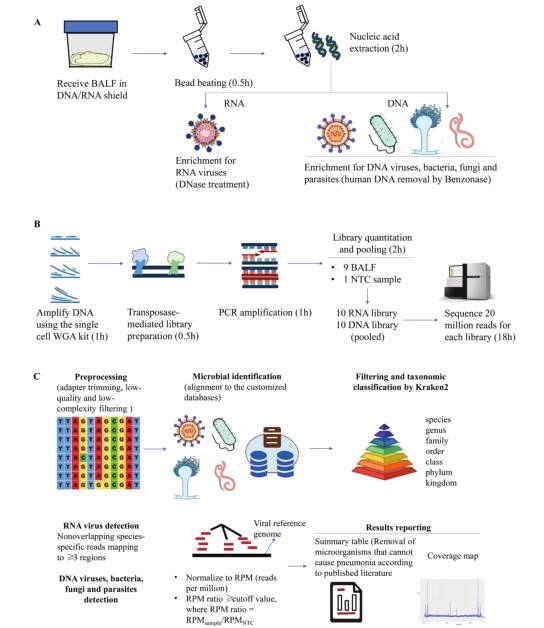 In-house mNGS assay workflow
In-house mNGS assay workflow
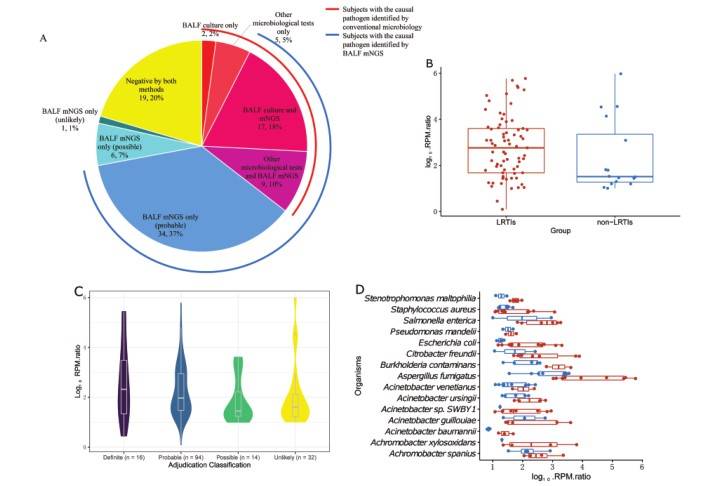 Clinical usability of mNGS for LRTI pathogen identification
Clinical usability of mNGS for LRTI pathogen identification
How to identify the pathogen in a short time is the key to early detection and treatment of respiratory tract infection. Metagenomic next-generation sequencing (mNGS) has a wide coverage and a large amount of information for the detection of pathogens of respiratory tract infections, which plays a very important role in the early detection of respiratory tract pathogens, especially in the rapid diagnosis of pathogens of new infectious diseases. In the future, it has broad application prospects in the future and will help us mine more information about respiratory tract infections.
References:
- Miao, Qing et al. "Evaluation of respiratory samples in etiology diagnosis and microbiome characterization by metagenomic sequencing." Respiratory research vol. 23,1 345. 14 Dec. 2022
- Chen, Hongbin et al. "Clinical Utility of In-house Metagenomic Next-generation Sequencing for the Diagnosis of Lower Respiratory Tract Infections and Analysis of the Host Immune Response." Clinical infectious diseases: an official publication of the Infectious Diseases Society of America vol. 71, Suppl 4 (2020): S416-S426.
- Yang, Lei et al. "Metagenomic Next-Generation Sequencing for Pulmonary Fungal Infection Diagnosis: Lung Biopsy versus Bronchoalveolar Lavage Fluid." Infection and drug resistance vol. 14 4333-4359. 20 Oct. 2021.


 Sample Submission Guidelines
Sample Submission Guidelines
