Application of Whole Plasmid Sequencing in Drug Resistance Genes of Klebsiella pneumoniae
Obtain the whole genome sequence of the plasmid through high-throughput sequencing, for predicting and annotating all genes in the genome, and finding genes related to phenotype, such as drug resistance genes, heavy metal resistance genes, and environmental adaptability genes. The "candidate" drug resistance gene proved by drug resistance genome research has resistance in the heterologous host, but it cannot prove that the gene still has resistance in its natural reservoir; On the other hand, these drug-resistant genes may undergo horizontal transfer between different bacteria in the form of plasmids, resulting in contamination of drug-resistant genes. Whole Plasmid Sequencing technology can screen out "candidate" drug resistance genes and annotate, which combines with drug resistance functional verification experiments can help us find the drug resistance mechanism as soon as possible and take corresponding countermeasures.
Plasmid Whole Genome Sequencing for Analyzing Drug Resistance Mechanisms
Klebsiella pneumoniae is one of the leading causes of hospital- and community-acquired infections, manifesting as pneumonia, urinary tract infections, septicaemia and abscesses. In recent years, the rapid dissemination of carbapenem-resistant K.pneumoniae (CRKP), a critical priority pathogen listed by WHO and has become a global threat to human health due to high morbidity and mortality. Researchers investigated the epidemiological characteristics of resistance and virulence factors of carbapenem resistant Klebsiella pneumoniae (CRKP), which isolated in pediatric patients in Shanghai. Through high-throughput sequencing, it was found that the isolate had a 5508387 bp ring chromosome and two plasmids pNDM-IMP-1 (347317 bp) and pNDM-IMP-2 (144371 bp). The strain was identified as ST3936 and KL30 by MLST and Kaptive typing. In addition, the strain carries multiple resistance genes, such as aminoglycosides, fluoroquinolones, carbapenems, and other β- Lactam drugs. The carbapenemase encoding genes blaNDM-1 (2 copies) and blaIMP-4 are co located on the plasmid pNDM-IMP-1, which is a 347.3 kb unclassifiable plasmid, containing most of the resistance genes except for oqxAB and blaLEN17. Comparing and analyzing the chromosomes of SHET-01 and Klebsiella mutabilis DX120E found that their genomic content was highly similar, confirming that SHET-01 belongs to Klebsiella mutabilis. Comparative genomics analysis found that pNDM - IMP - 1 and pKP1814 - 1 have similar plasmid skeletons and contain highly conserved plasmid functional genes (such as replicon gene repA and stability gene telB - stabD, etc.), conjugation transfer gene cluster, insertion elements, drug resistance determinants (such as blaIMP-4 and blaSFO) and other functional bases. This is the first report on the communist IMP-4 and NDM-1 carbapenemases in highly viscous Klebsiella pneumoniae. The multidrug-resistant hybrid plasmid pNDM-IMP-1 may originate from Klebsiella pneumoniae, and the ISCR1 element may contribute to the aggregation of blaNDM-1 and blaIMP-4 in this plasmid of Klebsiella pneumoniae strains. Long term monitoring of these strains can effectively prevent their outbreak and transmission.
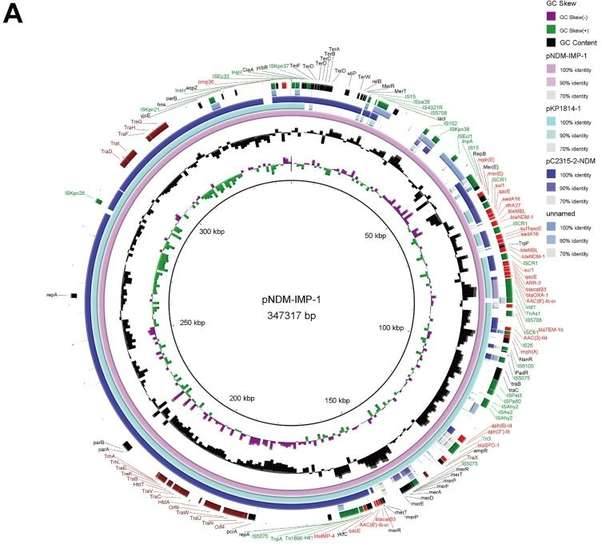 Comparative genomics analysis of plasmid and genetic structure
Comparative genomics analysis of plasmid and genetic structure
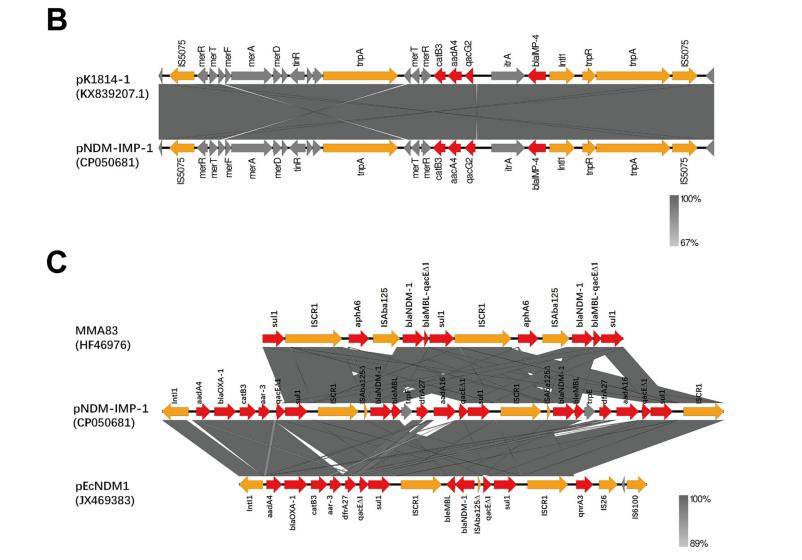 Plasmid Genome Circle Map and Plasmid Structure Genetic Background Map
Plasmid Genome Circle Map and Plasmid Structure Genetic Background Map
Whole genome sequencing for identification of Superplasmid
The emergence of carbapenem resistant HvKP (CR HvKP) and ESBL producing HvKP strains is mainly due to their simultaneous acquisition of high virulence and multidrug-resistant (MDR) genes (especially ESBL and carbapenemase), which are mainly mediated by plasmids. In previous reports, virulence and drug resistance genes were mostly located on different plasmids, but in recent years, plasmids contain both virulence and MDR genes have also begun to be reported. The emergence of this plasmid further accelerates the synchronous transmission of high virulence and MDR genes and brings more serious impacts on public health. Through the analysis of 239 strains of bloodstream infected pulmonary tuberculosis, we identified a clinical strain ST11-K64 CR-HvKP carrying a "super plasmid", revealed in detail its genomic structure and phenotypic characteristics. Usually, it has the following characteristics: (1) a single plasmid that coharbors hypervirulence and MDR genes, (2) a plasmid that harbors complete conjugative elements that guarantee self-transmissibility, (4) a plasmid that is stable and conserved, and (3) a plasmid with no fitness cost to the host strain. This self-transmissible superplasmid, which simultaneously carries hypervirulence and MDR genes, greatly enhances the challenges to clinical prevention and control and anti-infection treatment. Thus, active surveillance of this type of superplasmid is needed to prevent these efficient resistance/virulence plasmids from disseminating in hospital settings.
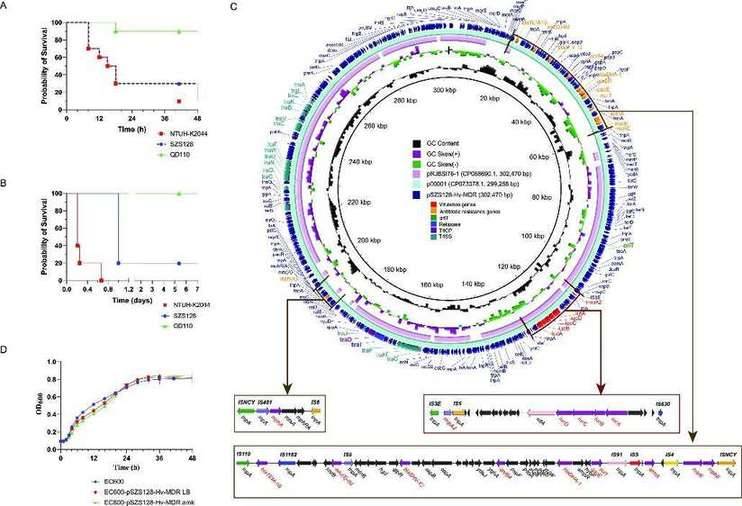 Virulence phenotype of CR-HvKP strain SZS128 and features of the superplasmid pSZS1280-Hv-MDR
Virulence phenotype of CR-HvKP strain SZS128 and features of the superplasmid pSZS1280-Hv-MDR
Antibiotic resistance is a major public health threat. Monitoring and understanding the prevalence, mechanism and spread of antibiotic resistance is the focus of individual patient care and overall infection control strategies. Whole plasmid sequencing has expanded our ability to detect drug-resistant genes in Klebsiella pneumoniae,which contributes to develop personalized treatment plans and provide convenience for the future use of conventional sequence based personalized medicine. This will also simplify the monitoring of antibiotic resistance.
References:
- Wang, Bingjie et al. "Genetic Characteristics and Microbiological Profile of Hypermucoviscous Multidrug-Resistant Klebsiella variicola Coproducing IMP-4 and NDM-1 Carbapenemases." Microbiology spectrum vol. 10,1 (2022): e0158121.
- Jia, Xinmiao et al. "Emergence of a Superplasmid Coharboring Hypervirulence and Multidrug Resistance Genes in Klebsiella pneumoniae Poses New Challenges to Public Health." Microbiology spectrum vol. 10,6 (2022): e0263422.


 Sample Submission Guidelines
Sample Submission Guidelines
