Integrating m6A Methylation and Single-Cell Analyses for Unraveling Cellular Heterogeneity
Single-cell sequencing enables us to dissect the intricate landscape of cellular diversity across various cell samples. This encompasses variations in cell population composition, shifts in cell proportions, and distinctions in gene expression profiles among different cell types and samples. With single-cell data, we gain the ability to pinpoint the specific influence of gene expression changes within individual cell types on the entire system. Nevertheless, the intricacies of how these gene expression alterations occur and the underlying regulatory factors remain elusive within single-cell datasets.
m6A methylation, a pervasive RNA modification, plays a pivotal role in modulating RNA stability, localization, translation, splicing, and transportation. m6A methylation sequencing data provides a lens through which we can comprehend the mechanisms governing gene expression changes from an RNA methylation perspective, shedding light on the intricacies of the gene expression modulation process.
The amalgamation of single-cell data with m6A methylation sequencing data offers a more comprehensive view of the regulatory mechanisms governing biological processes, enabling a deeper understanding of cellular heterogeneity and gene expression dynamics.
YTHDF2 Boosts Antitumor Immunity via TAM Reprogramming
In this study, the authors investigated the role of YTHDF2 in coordinating tumor-associated macrophage (TAM) reprogramming and its impact on anti-tumor immunity through CD8+ T cells. The research uncovered compelling insights into the interplay between YTHDF2, TAMs, and the immune response in various cancer types.
- Elevated YTHDF2 Expression in Multiple Cancer Types
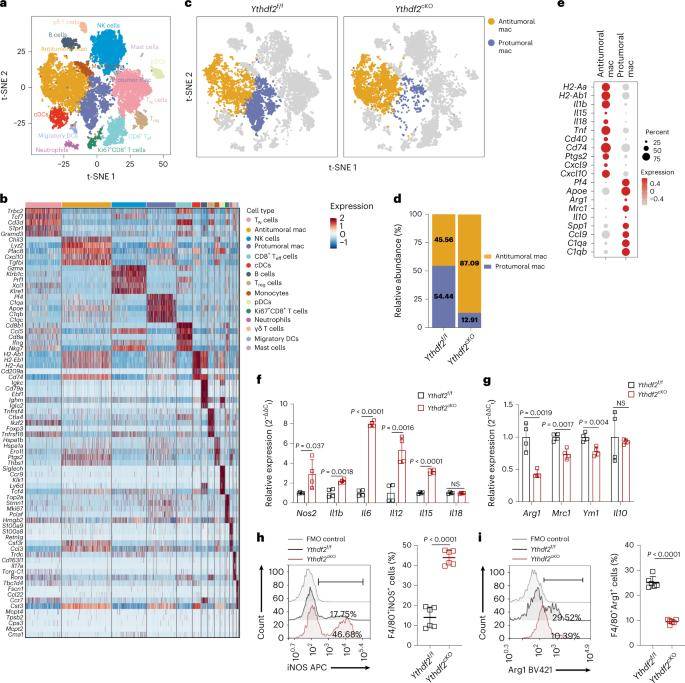
The authors initially examined the TCGA public database and observed that YTHDF2 exhibited notably high expression in tumor tissues of breast cancer (BRCA), colon adenocarcinoma (COAD), glioblastoma (GBM), rectal adenocarcinoma (READ), and cutaneous melanoma (SKCM). Importantly, they found a negative correlation between YTHDF2 mRNA expression and immune scores, implying a potential link between YTHDF2 and immune response. Furthermore, scRNA-seq data from public databases confirmed the upregulation of YTHDF2 in various tumor samples, as compared to neighboring or normal tissues, suggesting a common trend of YTHDF2 overexpression in tumors. - YTHDF2 Deletion Enhances Antitumor Phenotype in TAMs
Using single-cell sequencing of B16-OVA tumors from both Ythdf2f/f and Ythdf2cKO mice, the authors discovered increased expression of antitumoral markers and reduced expression of protumoral markers in TAMs following YTHDF2 deletion. This observation was further validated through cell sorting, cell identification analysis, and differential abundance analysis of cellular taxa. Collectively, these findings indicate that YTHDF2 deletion induces an antitumor program in TAMs. - Improved Antigen Cross-Presentation and CD8+ T Cell Activation
Cell communication analysis demonstrated that YTHDF2-deficient macrophages exhibit enhanced antigen cross-presentation and activation of CD8+ T cells. This suggests that YTHDF2 plays a pivotal role in modulating the interaction between TAMs and T cells, potentially influencing the anti-tumor immune response. - Regulation of the IFN-γ-STAT1 Signaling Pathway
The authors conducted various analyses, including GSEA analysis, qPCR experiments, phosphorylation experiments, and functional assays, to pinpoint the key genes and pathways affected by YTHDF2 deletion. Their findings revealed that YTHDF2 deletion regulates the IFNγ-STAT1 signaling pathway in macrophages, which is associated with the induction of an anti-tumor phenotype. - m6A Methylation Modification and Stat1 mRNA Stability
The study delved into the molecular mechanisms underlying YTHDF2's role in TAM reprogramming. Through m6A-seq and RIP-seq sequencing, the authors identified that YTHDF2 deletion enhances the stability of Stat1 mRNA in macrophages via m6A methylation modification, shedding light on the mechanistic details of YTHDF2's influence.
 Cell communication analysis. (Ma et al., 2023)
Cell communication analysis. (Ma et al., 2023)
In summary, this study provides compelling evidence that the m6A-reading protein YTHDF2 reprograms TAMs by influencing the IFN-γ-STAT1 pathway. This reprogramming results in improved TAM antigen presentation, ultimately modulating CD8+ T cell-mediated antitumor immunity. These findings offer valuable insights into the complex interplay between immune cells and the tumor microenvironment and have potential implications for cancer therapy.
METTL14 in Tumor-Associated Macrophages: Impact on CD8+ T Cells
In this groundbreaking study, the authors employed a comprehensive approach to investigate the impact of METTL14 deficiency in tumor-associated macrophages on CD8+ T cell dysfunction and tumor growth. Through a series of integrated experiments, single-cell sequencing, functional enrichment analysis, and RNA-seq, they uncovered a novel regulatory mechanism involving m6A methylation. This mechanism revealed how the loss of the m6A methylase METTL14 in C1q+ macrophages leads to a decrease in m6A modification on the Ebi3 transcript and an increase in EBI3 expression in macrophages. This, in turn, triggers dysfunction in tumor-infiltrating CD8 T cells, with profound implications for cancer immunology.
- Characterization of Macrophage Subpopulations
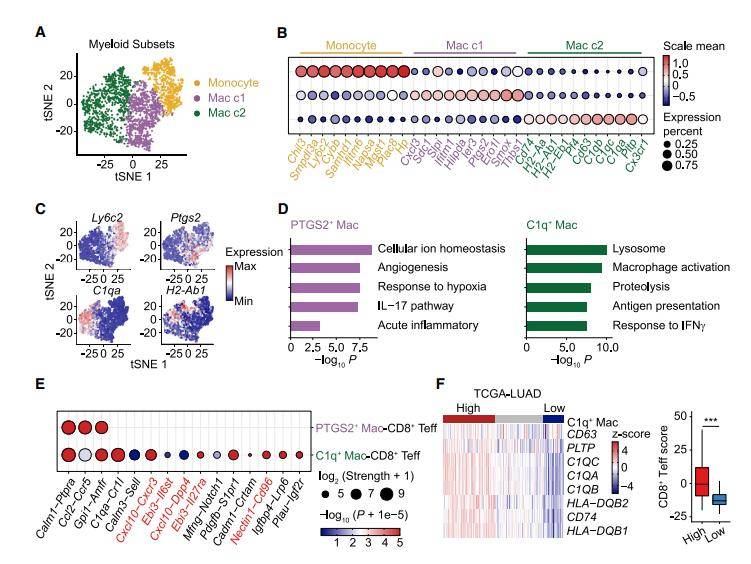
To initiate their investigation, the authors meticulously classified monocytes and macrophages into three distinct subpopulations using single-cell sequencing data. The Mac c1 subpopulation, referred to as PTGS2 macrophages, was identified as having potential roles in angiogenesis and hypoxia. In contrast, the Mac c2 subpopulation, known as C1q macrophages, exhibited features induced by interferon γ (IFN-γ), hinting at interactions with IFNγ-producing cells, particularly effector T cells. - Monocyte Differentiation Trajectories
The authors then focused on monocyte differentiation trajectories and observed the differentiation of tumor-infiltrating monocytes into either C1q or PTGS2 macrophages. Importantly, they identified transcription factors, such as Klf2 and Klf4 for M2 macrophage polarization, to be elevated along the C1q macrophage trajectory, while PTGS2 macrophages expressed Jun and Ets1. - Validation of RNA m6A Methylation in Macrophages
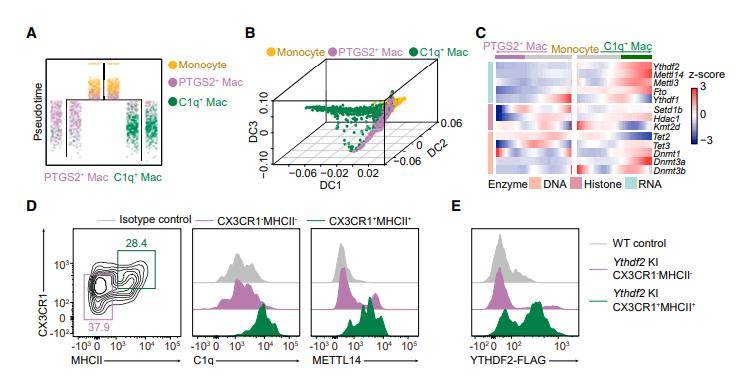
To validate the presence of RNA m6A methylation in macrophages, bulk RNA-seq analyses confirmed that genes representative of C1q macrophages and PTGS2 macrophages, as identified in single-cell RNA-seq analysis, were reproducible in bulk-sorted MHCII and MHCII subsets, respectively. Notably, C1q, METTL14, and YTHDF2 were predominantly expressed in CX3CR1MHCII macrophages, further supporting the relevance of these findings. - Impact of METTL14 Deficiency on Tumor Immunity
Subsequently, the authors explored the effects of METTL14 deficiency in macrophages in the context of tumor immunity. Their experiments involving mice with METTL14-specific knockout in macrophages revealed reduced anti-tumor capacity compared to wild-type mice. They also observed a significant decrease in the proportion of tumor-infiltrating CD8 T cells. Single-cell sequencing data demonstrated that METTL14 deficiency in macrophages led to a decline in infiltrating effector CD8 T cells and a rise in abnormally activated and dysfunctional intermediate-state CD8 T cells in the immune microenvironment. - Functional Validation and Mechanistic Insights
Functional validation experiments further illuminated that METTL14-deficient macrophages diminished the killing function of CD8 T cells and upregulated the expression of co-suppressive receptors. This indicated that METTL14-deficient macrophages disrupted the equilibrium of CD8 T cell differentiation, inhibiting effector CD8 T cell activation and contributing to CD8 T cell dysfunction. Integrating m6A-seq and RNA-seq data from wild-type and METTL14-deficient tumor-associated macrophages, the authors identified a significant reduction in m6A modifications on Ebi3 mRNA in the METTL14-deficient macrophages. Additionally, the levels of Ebi3 mRNA and protein expression were significantly increased. - Therapeutic Potential
To further probe the mechanistic underpinnings of these observations, the authors demonstrated that neutralizing EBI3 with an EBI3-neutralizing antibody restored CD8 T cell differentiation and enhanced their anti-tumor capabilities in conditional knockout mice. These results highlighted the role of METTL14-mediated methylation of Ebi3 in C1q macrophages as a pivotal switch in shaping the tumor microenvironment towards an immunosuppressive phenotype. Importantly, blocking the negative regulation of T cell function by Ebi3 effectively counteracted the immunosuppressive effects induced by the loss of METTL14 in tumor-associated macrophages.
 Characterization of tumor-associated macrophages by scRNA-seq. (Dong et al., 2021)
Characterization of tumor-associated macrophages by scRNA-seq. (Dong et al., 2021)
 C1q+ macrophage is distinguished by unique RNA m6A methylation. (Dong et al., 2021)
C1q+ macrophage is distinguished by unique RNA m6A methylation. (Dong et al., 2021)
References:
- Ma, Shoubao, et al. "YTHDF2 orchestrates tumor-associated macrophage reprogramming and controls antitumor immunity through CD8+ T cells." Nature immunology 24.2 (2023): 255-266.
- Dong, Lihui, et al. "The loss of RNA N6-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8+ T cell dysfunction and tumor growth." Cancer Cell 39.7 (2021): 945-957.


 Sample Submission Guidelines
Sample Submission Guidelines
