Human Leukocyte Antigen (HLA) Expression and Immune Modulation in Pan-Cancer Immunogenicity
Background
Human Leukocyte Antigen (HLA) molecules play a pivotal role in immune response modulation by presenting antigenic peptides to T cell receptors (TCRs). This interaction is crucial for initiating anti-tumor immune responses. However, the systematic role of HLA expression in pan-cancer immunogenicity and immunoediting mechanisms remains underexplored. In this case, researchers aimed to investigate the differential expression of HLA class I and class II genes across tumor types, oncogenic pathways, and immune subtypes.
Methods
The primary objective of this study was to comprehensively investigate the role of HLA-mediated mechanisms in tumor immunogenicity and escape across various tumor types. The study aimed to understand the differential expression of HLA class I and class II genes, their correlations with immune infiltration, their associations with oncogenic pathways and immune subtypes, and their potential implications for patient survival and immunotherapy response.
Researchers conducted a retrospective analysis of genomic and clinical data from 33 tumor types obtained from the Cancer Genome Atlas (TCGA) dataset. RNA sequencing data were used to quantify the expression levels of HLA class I and class II genes across different tumors. Immune subtypes were defined using established gene expression signatures. Oncogenic pathways were categorized based on molecular alterations. Machine learning techniques were employed to predict immune subtypes and stratify patient survival based on HLA gene expression patterns.
Data Collection and Preprocessing
- The Cancer Genome Atlas (TCGA) dataset was utilized, encompassing genomic and clinical data from multiple tumor types.
- RNA sequencing data were used to quantify HLA class I and class II gene expression levels across different tumor types and molecular subtypes.
- HLA supertypes and allelic diversity were characterized to assess the correlation between germline HLA heterozygosity and patient survival.
HLA Expression and Immune Infiltration
- Correlations between HLA class I and class II gene expression levels and immune infiltration were analyzed to determine the relationship between HLA expression and tumor immune response.
Differential Expression Across Oncogenic Pathways and Immune Subtypes
- HLA gene expression profiles were examined across different oncogenic pathways and immune subtypes to identify patterns of HLA expression associated with specific tumor characteristics.
Epigenetic Modifications and HLA-Mediated Escape
- DNA methylation at HLA genes and loss of heterozygosity (LOH) at HLA loci were investigated as potential mechanisms for HLA-mediated tumor escape strategies.
Machine Learning Model for Immune Subtype Prediction
- A machine learning model was trained using HLA gene expression data from the TCGA Skin Cutaneous Melanoma (SKCM) dataset to predict immune subtypes.
- The trained model was then applied to predict immune subtypes in an independent cohort of melanoma patients who had received immunotherapy.
Results
- HLA Expression and Immune Subtypes: They observed that HLA class I genes were consistently upregulated in tumors characterized as immunogenically "hot," indicating active anti-tumor immunity. In contrast, HLA class II genes were predominantly upregulated in an inflammatory immune subtype associated with the best prognosis. Notably, HLA class II genes were systematically downregulated in specific oncogenic pathways, suggesting a potential immune evasion mechanism.
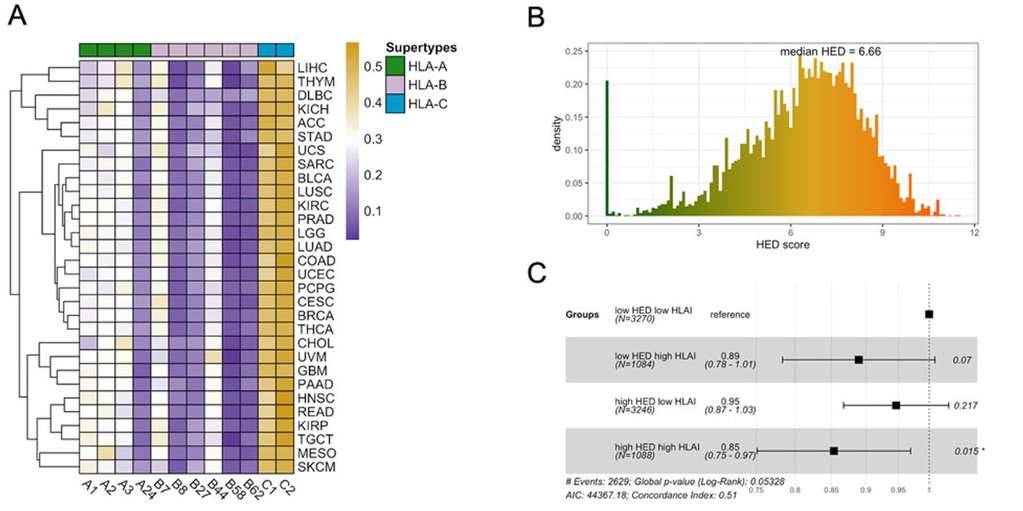 HLA Supertypes and Heterozygosity. (Gong et al., 2022)
HLA Supertypes and Heterozygosity. (Gong et al., 2022)
- HLA-Mediated Immune Escape: Among the immunogenically hot tumors, a subset displayed upregulation of HLA class I genes without concurrent upregulation of HLA class II genes. This observation suggested that these tumors might employ HLA-mediated escape strategies to evade immune detection, potentially leading to resistance against immune surveillance.
- Predictive Value of HLA Expression: By employing a machine learning model, they demonstrated that HLA gene expression data could accurately predict the immune subtypes of patients undergoing immune checkpoint blockade therapy. Furthermore, based on HLA expression patterns, they were able to stratify patient survival, indicating the potential clinical relevance of HLA-mediated immune modulation.
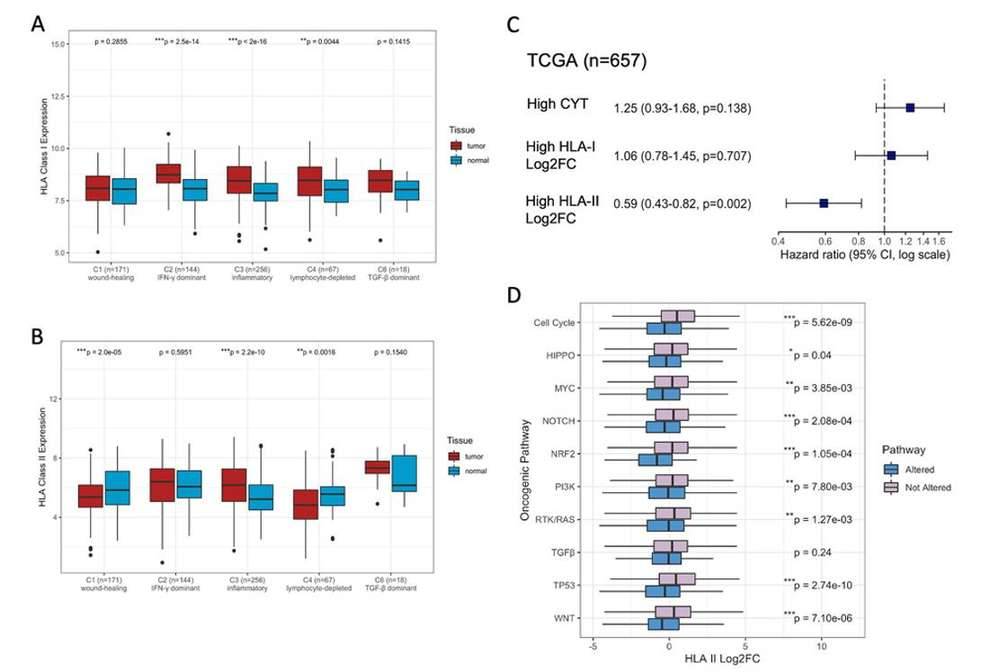 HLA Differential Expression Associated with Immune Subtypes and Oncogenic Pathways. (Gong et al., 2022)
HLA Differential Expression Associated with Immune Subtypes and Oncogenic Pathways. (Gong et al., 2022)
- Immune Infiltration and Prognosis: Interestingly, they found that tumors with the highest levels of immune infiltration did not necessarily exhibit the best prognosis. Instead, these tumors showed significantly higher levels of immune exhaustion, suggesting that excessive immune activity might lead to dysfunctional immune responses and compromised tumor control.
Conclusion
This case study provides comprehensive insights into the role of HLA expression in pan-cancer immunogenicity and immunoediting mechanisms. The findings highlight the distinct roles of HLA class I and class II genes in modulating anti-tumor immune responses, as well as their potential implications for immune evasion strategies. The predictive power of HLA gene expression for immune subtypes and patient survival underscores its clinical relevance in guiding personalized immunotherapeutic strategies. Moreover, the association between immune infiltration, immune exhaustion, and prognosis emphasizes the delicate balance required for effective anti-tumor immunity.
Reference:
- Gong, Xutong, and Rachel Karchin. "Pan-cancer HLA gene-mediated tumor immunogenicity and immune evasion." Molecular Cancer Research 20.8 (2022): 1272-1283.


 Sample Submission Guidelines
Sample Submission Guidelines
