EWAS Reveals Methylation Sites Associated with Multiple Common Diseases
Epigenome-wide association studies (EWAS) offer an effective approach to explore the genetic basis of complex diseases and investigate the relationship between CpG methylation and associated health outcomes. Common disease EWAS typically involve whole-blood DNA analysis and can be classified into prevalence and incidence analyses. Previous studies had limited sample sizes, often below 1000 individuals, and focused on specific conditions, thus restricting the identification of methylation sites associated with disease states. Consequently, there is a pressing need for large-scale EWAS that encompass multiple disease states within a single population, providing insights into the relevance of blood DNA methylation as a peripheral marker for common diseases.
A groundbreaking study titled "Blood-Based Epigenome-Wide Analysis Reveals Methylation Sites Linked to Multiple Common Diseases" was recently published in PLOS Medicine. This study focuses on DNA methylation, the foremost and extensively studied epigenetic modification of genes. DNA methylation occurs mainly at cytosine-phosphate-guanine dinucleotide (CpG) sites and plays a vital role in various aspects of cellular function, chromosome structure, embryonic development, aging, disease onset, and the interaction between the environment and the genome. Understanding the mechanisms and functions of DNA methylation holds immense potential for advancing human health and disease research.
In response to this need, the research team conducted a comprehensive EWAS analysis on the Generation Scotland (GS) cohort, which comprised 18,413 individuals with available DNA methylation data. Their study examined the association between methylation differences at CpG sites and the prevalence of 14 disease states, as well as the incidence of 19 disease states. The impressive results identified more than 100 CpG methylation sites associated with the prevalence of four disease states (breast cancer, chronic kidney disease, ischemic heart disease, and type 2 diabetes mellitus) and the incidence of two disease states (chronic obstructive pulmonary disease and type 2 diabetes mellitus).
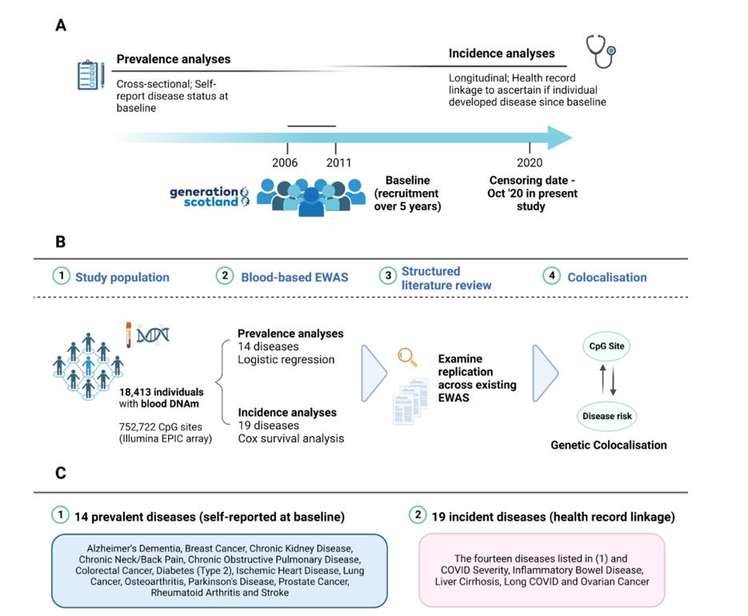 Study design for epigenome-wide analyses on prevalent and incident disease states in Generation Scotland. (Hillary et al., 2023)
Study design for epigenome-wide analyses on prevalent and incident disease states in Generation Scotland. (Hillary et al., 2023)
Epigenome-Wide Analysis of Prevalence of 14 Diseases
The prevalence of 14 diseases was analyzed through an epigenome-wide approach. The research team examined DNA methylation at 752,722 CpG loci in whole blood samples from 18,413 participants. They integrated this DNA methylation data with self-reported disease information from the baseline study to conduct an Epigenome-Wide Association Study (EWAS) on these 14 common disease states.
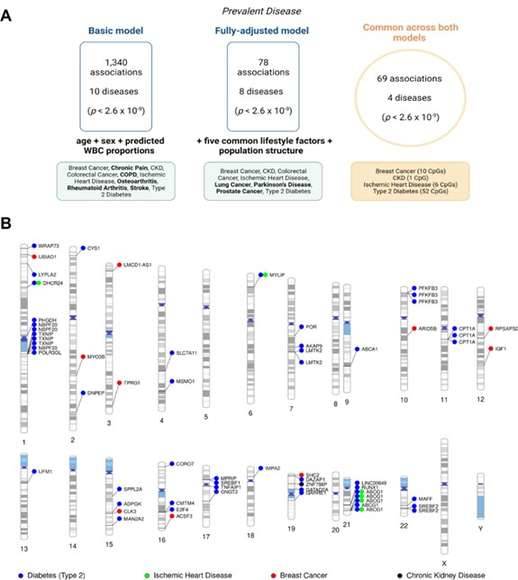 Epigenome-wide association studies on 14 prevalent disease states in Generation Scotland. (Hillary et al., 2023)
Epigenome-wide association studies on 14 prevalent disease states in Generation Scotland. (Hillary et al., 2023)
Initially, using a base model, the researchers identified 1,340 significant associations between blood CpG methylation and 10 diseases. Remarkably, more than 90% of these associations were linked to type 2 diabetes, chronic obstructive pulmonary disease (COPD), and chronic pain.
To account for potential confounding factors, the research team then fully adjusted the model by considering the effects of five common lifestyle risk factors and demographics: alcohol consumption, body mass index, poverty level, smoking, and years of education. In the fully adjusted model, they found 78 associations with 8 disease states, out of which 69 associations overlapped with the associations found in the basic model. These 69 associations were distributed across 4 disease states: chronic kidney disease, ischemic heart disease, breast cancer, and type 2 diabetes.
Of particular interest, the analysis revealed that participants with a self-reported history of breast cancer showed hypomethylation of cg06072257 and cg06123699, which are located near UBIAD1 and TPRG1 on chromosomes 1 and 3, respectively. Additionally, CpG methylations annotated as ABCG1, DHCR24, and MYLIP were more prevalent in individuals with ischemic heart disease and type 2 diabetes.
Exploring 19 Disease Outbreaks Through Epigenome-wide Analysis
Using electronic health record linkage and long-term follow-up monitoring, a group of researchers investigated 19 emergent disease states. They conducted an Epigenome-wide Association Study (EWAS) to determine whether CpG methylation sites identified during the baseline study could be linked to the future onset of these 19 diseases.
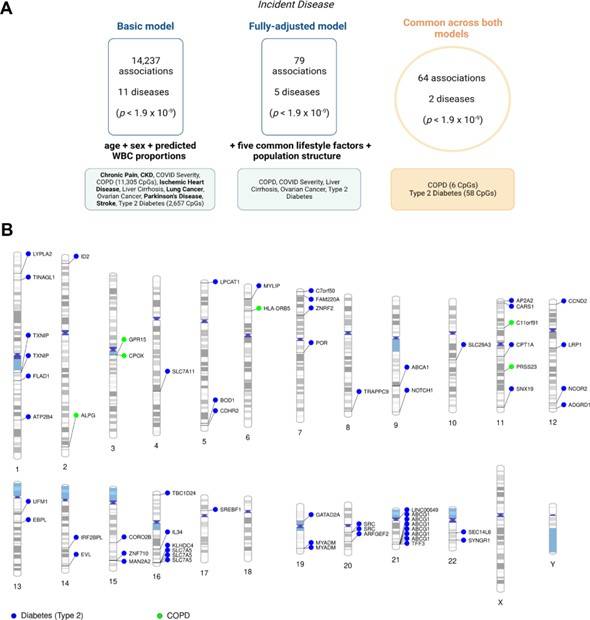 Epigenome-wide association studies on 19 incident disease states in Generation Scotland. (Hillary et al., 2023)
Epigenome-wide association studies on 19 incident disease states in Generation Scotland. (Hillary et al., 2023)
In the initial model, the team discovered 14,237 associations between CpG methylation and the incidence of 11 disease states. Notably, 79.4% of these associations were linked to COPD, while 18.7% were related to type 2 diabetes.
Upon employing a fully adjusted model, the researchers identified 79 unique associations across 5 disease states. Surprisingly, only 64 associations remained consistent between the basic and fully adjusted models, specifically involving COPD and type 2 diabetes.
Among the CpG-annotated genes associated with COPD were ALPG, C11orf91, CPOX, GPR15, HLA-DRB5, and PRSS23. On the other hand, the CpG-annotated genes linked to type 2 diabetes included ABCA1, ABCG1, CPT1A, SREBF1, SLC7A11, SLC7A5, and TXNIP.
To delve deeper, the team examined the fully adjusted model and tested the influence of five common lifestyle risk factors on the aforementioned associations. They found that each covariate had a different attenuating effect on the effect sizes, ranging from 5.5% for body mass index to 63.1% for smoking. These effects varied based on the unique risk profiles of different disease states.
Leveraging one of the largest methylation datasets worldwide, the research team embarked on a series of EWAS to explore the prevalence and incidence of multiple disease states. To augment their findings, they conducted a comprehensive review of existing literature on a large scale. Through meticulous comparison between this study's results and the gathered data, the team unveiled 58 previously unknown associations with the prevalence of three disease states—breast cancer, ischemic heart disease, and type 2 diabetes. Additionally, they discovered 56 novel connections between CpG methylation sites and the incidence of two disease states—COPD and type 2 diabetes—that were independent of common lifestyle risk factors. The implications of this study propose that blood DNA methylation could serve as a valuable peripheral biomarker for several widespread disease conditions, including breast cancer, cardiopulmonary disease, and type 2 diabetes.
References:
- Hillary, Robert F., et al. "Blood-based epigenome-wide analyses of 19 common disease states: A longitudinal, population-based linked cohort study of 18,413 Scottish individuals." PLoS Medicine 20.7 (2023): e1004247.


 Sample Submission Guidelines
Sample Submission Guidelines
