Comprehensive Mapping of Regulatory Elements in the Chicken Genome
The chicken, the pioneer among agricultural animals to have its genome sequenced, stands as a cornerstone of domestication. As one of the most extensively studied bird species, it provides valuable insights into the evolutionary connections between mammals and avian species. Beyond its status in the agricultural realm, the chicken serves as a crucial model organism in various scientific disciplines, including evolutionary and developmental biology, immunology, and epigenetic gene regulation. The endeavor to annotate functional elements in livestock genomes plays a pivotal role in deciphering the molecular underpinnings of economically significant and intricate traits such as growth, reproduction, and disease resistance. This endeavor, especially in terms of comprehensive annotation of non-coding regions, is an urgent necessity. In analogy to the Encyclopedia of DNA Elements (ENCODE) and the Epigenome Roadmap projects, the Functional Annotation of Animal Genomes (FAANG) consortium is making remarkable strides in enhancing genome annotation for livestock and poultry.
Nonetheless, the current annotation of regulatory elements remains confined to a limited set of tissues, especially in the chicken. This underscores the pressing need for a more exhaustive regulatory element mapping. The comprehensive mapping of regulatory elements in the chicken genome promises to facilitate the identification of potential causal variant sites for economically pivotal traits. This, in turn, accelerates the breeding process and further enriches the utility of chickens as research subjects in life sciences.
This study stands as a pioneering effort in the construction of a regulatory element map across various tissues, integrating data from 377 distinct epigenomic, transcriptomic, and chromatin conformation datasets derived from 23 major adult chicken tissues. It delves into the nuanced, tissue-specific functions of regulatory elements and anticipates interactions between enhancers and their target genes, even spotlighting the concept of super-enhancers.
In illuminating the functional facets of these elements, this research contributes to our comprehension of diverse aspects such as chicken domestication, monogenic traits, and the intricate regulation of economically significant traits.
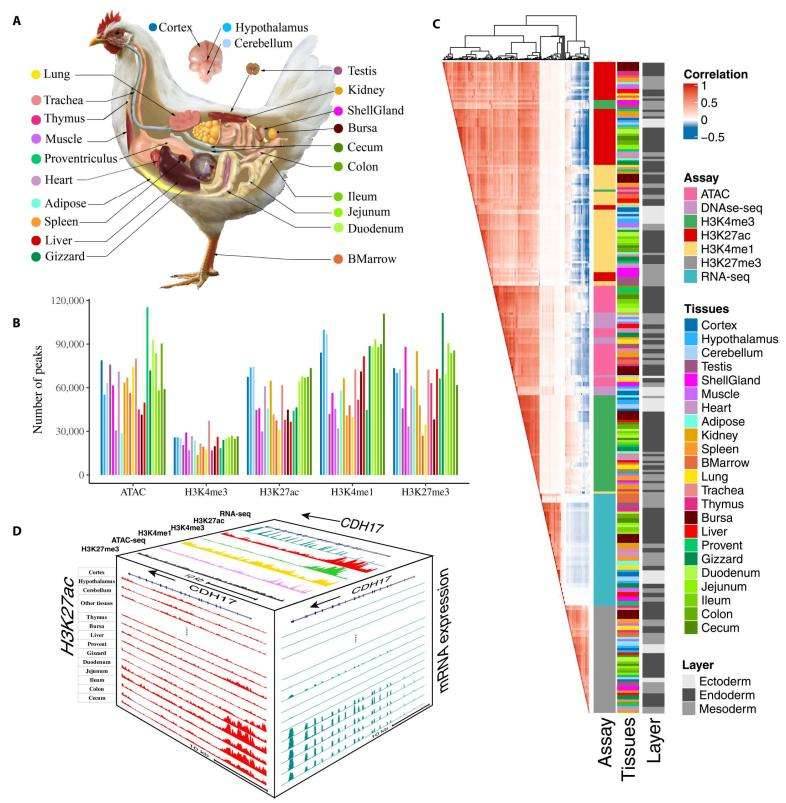 The summary of chicken epigenomic atlas. (Pan et al., 2023)
The summary of chicken epigenomic atlas. (Pan et al., 2023)
Tissue-Specific Gene Regulation in Chickens
In this study, 377 genome-wide datasets were generated from 23 major chicken tissues using a combination of advanced technologies, including ChIP-seq (for histone H3K4me3, H3K4me1, H3K27ac, H3K27me3, and CTCF), ATAC-seq, DNase-seq, RRBS, RNA-seq, and Hi-C. These datasets were meticulously assessed in the context of diverse histological assays and tissues. They shed light on the intricate relationship between regulatory elements and gene expression.
As a notable example, the datasets revealed that CDH17 exhibited specific and pronounced expression in intestinal tissues compared to other tissue types. Furthermore, it was conspicuously enriched near the transcription start site (TSS) within the H3K27ac modification, mirroring the results of other active regulatory markers, in contrast to inhibitory regulatory modification markers. These findings collectively emphasize how they, the genes expressed in a tissue-specific manner in chickens, are cohesively regulated by different epigenetic markers.
Deciphering Chromatin States
Subsequently, by amalgamating data from all five epigenetic marks across 23 distinct tissues, the authors of ChromHMM successfully predicted 15 unique chromatin states. As anticipated, these states exhibited differential enrichment patterns: promoters predominated in the vicinity of the transcription start sites (TSS), 5' end non-coding regions, and CpG islands. Both active promoters and enhancers were significantly overrepresented in the TSS and expressed genes when compared to repressed genes.
Among these chromatin states, enhancer activity demonstrated the greatest dynamism across tissues, while promoter activity remained remarkably conserved. Promoters exhibited higher conservation levels than enhancers and repression regions at the DNA sequence level. The authors also delved into the intricate interplay between regulatory elements and gene expression in proximity to specific evolutionary breakpoint regions in the chicken genome, as well as the intricate relationship between DNA methylation and regulatory elements.
Their investigation particularly honed in on the interactions between enhancers and specific target genes, illustrating their role in regulating gene expression. Notably, genes associated with an abundance of enhancers displayed elevated levels of gene expression, diminished tissue-specific expression patterns, and heightened sequence-level conservation in comparison to genes with fewer associated enhancers. To exemplify these concepts, the authors employed the ovalbumin-associated protein (OVAL) gene locus to elucidate how regulatory elements orchestrate gene expression across a spectrum of tissues.
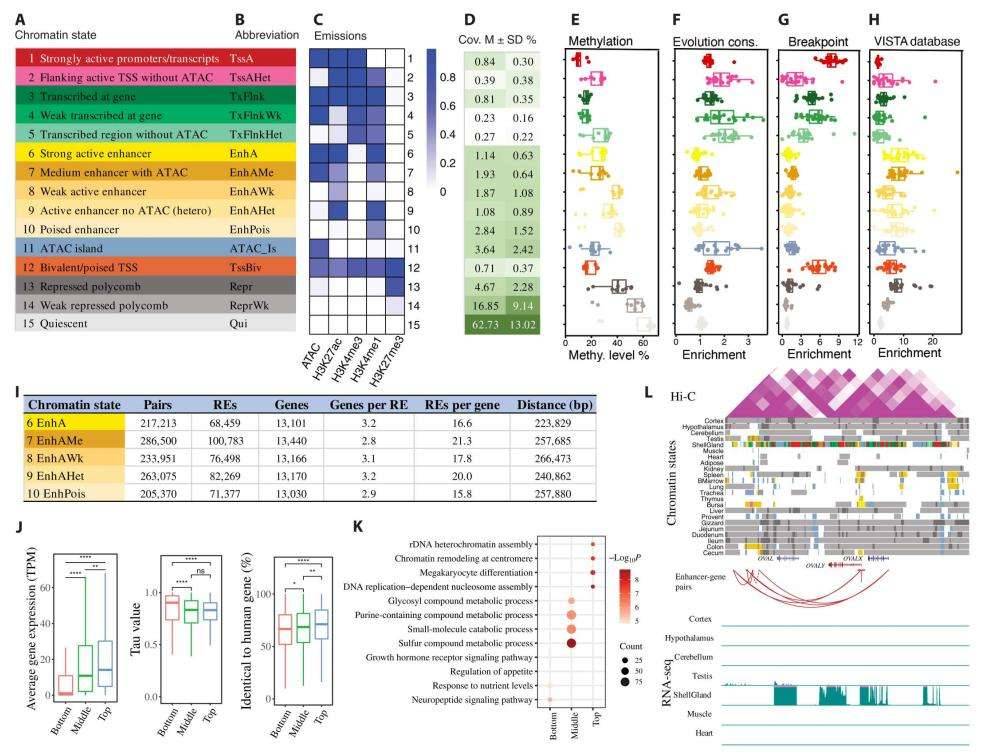 Discovery and characterization of chromatin states and enhancer-gene interactions in chicken. (Pan et al., 2023)
Discovery and characterization of chromatin states and enhancer-gene interactions in chicken. (Pan et al., 2023)
The Role of Regulatory Elements in Chicken Genetics and Evolution
In this study, they explore the application of elevated genome annotation in chicken biology research. When examining single-gene traits, they harness causal variants residing in the non-coding regions of seven specific traits included in the database. Remarkably, their findings reveal that six of these traits (constituting 75%) are intricately linked to one or more meticulously organized active regulatory elements. Notable examples include trait variant loci associated with polydactyly (SNP) and the coloration of chicken plumage (deletion), both of which were pinpointed within these regulatory domains.
Furthermore, their investigation encompasses a comprehensive analysis of complex traits, specifically examining GWAS summary statistics for 44 economically significant traits in chickens, including parameters like growth and egg production. Notably, they uncovered a heightened prevalence of active regulatory elements linked to GWAS signals.
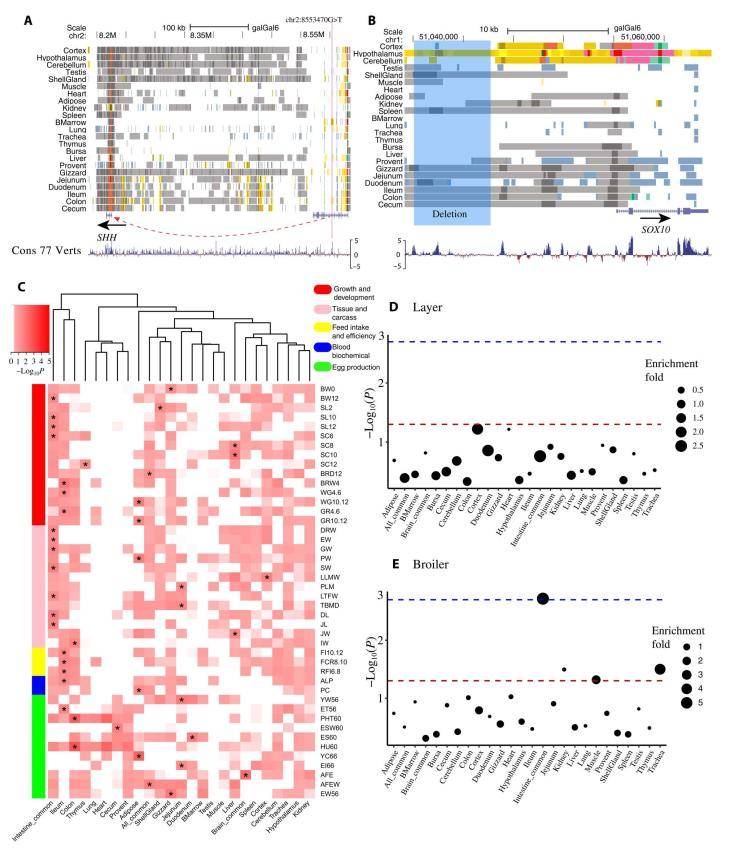 The regulatory elements involved in chicken monogenic traits, complex traits, and selection signatures. (Pan et al., 2023)
The regulatory elements involved in chicken monogenic traits, complex traits, and selection signatures. (Pan et al., 2023)
Their study also delves into the intriguing relationship between domestication and regulatory elements. It is evident from their research that domestication appears to be driven by tissue-specific enhancer regulation. This, when combined with previously documented findings concerning the selective processes during domestication, notably in the transition from red junglefowl to domestic chickens and the differentiation between modern broilers and laying hens, highlights a compelling interplay of factors. These observations suggest that their mapping of regulatory elements furnishes valuable insights into the characterization of candidate loci. This, in turn, holds the promise of enriching their comprehension of the molecular mechanisms underpinning the evolution and selection of both single genes and complex traits, as well as adaptations within the domain of chickens.
Reference:
- Pan, Zhangyuan, et al. "An atlas of regulatory elements in chicken: A resource for chicken genetics and genomics." Science Advances 9.18 (2023): eade1204.


 Sample Submission Guidelines
Sample Submission Guidelines
