Transcriptomic Signatures Across 40 Common Human Brain Diseases
Brain disorders are very diverse and commonly include cerebrovascular, neurodegenerative, motor-related, psychiatric disorders, developmental and congenital disorders, substance abuse disorders, brain tumors, and a range of other brain-related disorders. The etiology of brain disorders and their genetics are complex and widely studied, but phenotypic classification of brain disorders remains challenging. Except for inherited disorders caused by single gene mutations, most brain disorders manifest complex genetic-environmental interactions through the interaction of the brain transcriptome and its regulatory networks. Therefore, genetic analysis of brain diseases provides an important way to understand the molecular relationships.
Recently, an article entitled "A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases" was published in Plos Biology. The team analyzed gene expression data from the Allen Human Brain Atlas and identified five major transcriptional patterns representing tumor-associated, neurodegenerative, psychiatric and substance abuse, and two mixed groups of diseases affecting the basal ganglia and hypothalamus through the analysis of 40 common human brain diseases. In addition, localization by homologous cell types between mice and humans revealed that most disease-risk genes function in common cell types while having species-specific expression and maintaining similar phenotypic classification within species.
In conclusion, this study describes the structural and cellular transcriptome relationships of disease-risk genes in the adult brain, providing a molecular strategy for disease classification and comparison that is expected to provide important insights for the development of research methods in brain pathophysiology.
Neuroanatomical and Transcriptomic Analysis of Brain Diseases
The study involved 104 structures in the cerebral cortex, hippocampus, amygdala, basal ganglia, ependymal bodies, thalamus, ventral thalamus, hypothalamus, midbrain, cerebellum, pons, pontine nuclei, medulla, ventricles and white matter. By normalizing the average expression of 40 disease-associated genes involved in these structures, the team obtained transcriptomic expression profiles of diseases, and whole-brain transcriptomic clustering based on this identified a total of five major anatomical disease groups (ADG 1-ADG 5), including tumor-related (ADG 1), neurodegenerative (ADG 2), psychiatric, substance abuse and movement disorders (ADG 3) , associated with hypothalamic function, without developmental, psychiatric or tumor-related disorders (ADG 4), and basal ganglia-related disorders (ADG 5).
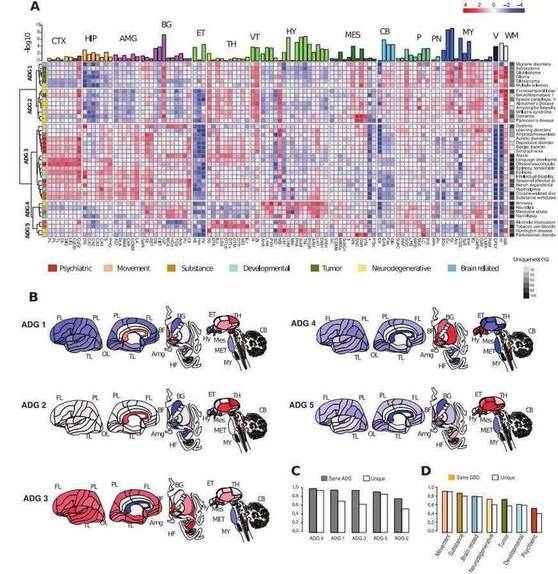 Transcriptome analysis of brain diseases. (Zeighami et al., 2023)
Transcriptome analysis of brain diseases. (Zeighami et al., 2023)
Typical Expression Patterns of Brain Diseases
Researchers have described the co-expression patterns and major constituent cell types of brain diseases by mapping disease genes to typical expression patterns. The M1 module has strong telencephalic expression in the hippocampus, particularly in the dentate gyrus, and representative genes include GRIA2 and DLG3, with mutations in GRIA2 associated with mental and neurodevelopmental disorders. The dopamine transporter gene SLC6A3, a candidate risk gene for dopamine or other toxins in dopamine neurons, and aldehyde dehydrogenase-1 (ALDH1A1), whose polymorphisms are associated with alcohol use disorders, both map to module M12.
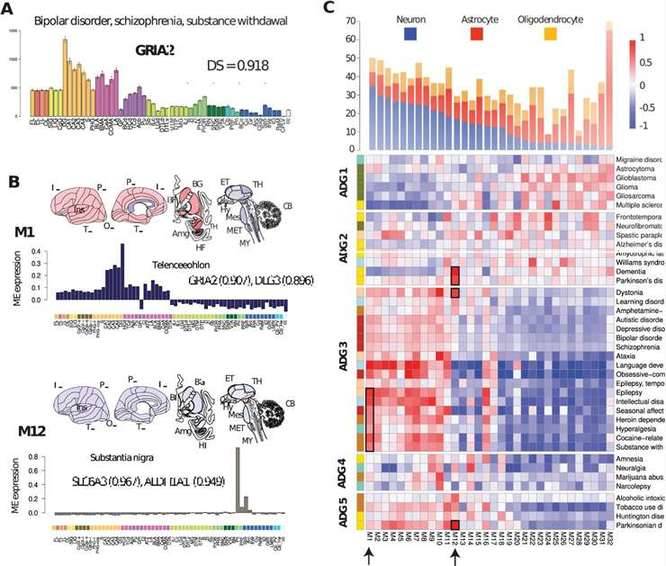 Reproducible transcription patterns in human brain diseases. (Zeighami et al., 2023)
Reproducible transcription patterns in human brain diseases. (Zeighami et al., 2023)
Comparison of Mouse and Human Brain Diseases
By performing a comparative analysis of homologous cell types in mouse and human brain, the team examined the conservation of disease-based cellular architecture between mice and humans, and the results indicate that cellular architecture is largely conserved between cortical regions and species. In addition, using expression-weighted cell type enrichment analysis (EWCE) to assess all genes in the disease simultaneously, the distribution of cell type expression can be determined and the characteristics of active enriched cell types in the disease can be characterized. The correlation of EWCE values between mice and humans reflects broad conserved expression patterns with minimal differences in global EWCE distribution. Overall, homologous localization of mouse and human cell types revealed that mouse and human disease risk genes function in homologous cell types with distinct species-specific expression.
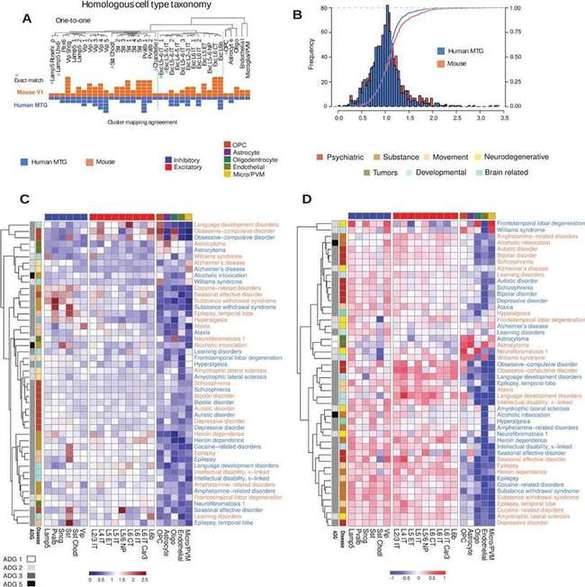 Disease-based cell type expression in mouse and human. (Zeighami et al., 2023)
Disease-based cell type expression in mouse and human. (Zeighami et al., 2023)
This study describes the structural and brain-wide transcriptomic relationships of risk genes for common brain diseases in the adult brain and proposes a brain-wide molecular signature of common brain diseases from a neuroanatomical structural perspective. Although the study does not currently allow precise identification of disease-specific gene expression changes, it still provides a new approach to characterize the cellular basis of disease.
Reference:
- Zeighami, Yashar, et al. "A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases." Plos Biology 21.4 (2023): e3002058.


 Sample Submission Guidelines
Sample Submission Guidelines
