Targeted RNA Sequencing: Unraveling the Detection Principles, Workflow and Diverse Applications
RNA-seq, a transcriptome sequencing analysis technology enabled by high - throughput sequencing, initially focused on studying the expression levels and differences of mRNA, small RNA, and non - coding RNA. Over the past decade, it has witnessed remarkable development and is now extensively applied in scenarios such as transcription variation analysis, gene fusion detection, and alternative splicing identification. Targeted RNA - seq, in contrast, zeroes in on the analysis of specific transcripts. Similar to standard RNA - seq, it utilizes targeted enrichment methods for gene expression evaluation, RNA species analysis, gene fusion and mutation detection. However, it outperforms standard RNA - seq in terms of sensitivity, dynamic range, cost - effectiveness, and throughput.
What is Targeted RNA-seq
Targeted RNA sequencing represents a high - throughput sequencing technique. Its objective is to analyze specific RNA transcripts to detect phenomena like gene fusions, mutations, splicing variations, and phenotype - specific expressions. By selectively amplifying the cDNA of the target region, it reduces sequencing costs and time compared to whole - transcriptome RNA - seq, while enhancing the sensitivity and specificity of experiments. The key features of targeted RNA - seq are as follows:
Target region selectivity: Targeted RNA-seq selectively amplifies the target RNA region through capture probes (such as biotin-labeled oligonucleotide probes) or specific primers. This approach allows for the in-depth analysis of specific genes, transcripts, or gene families.
Sensitivity and specificity: Owing to its focus on specific regions, targeted RNA-seq exhibits higher sensitivity in detecting low - abundance genes, rare fusion genes, or specific splicing variations. For instance, in acute myeloid leukemia (AML), it can uncover gene fusions that are elusive to traditional detection methods.
Gene fusion detection: This technique is widely employed in cancer research, particularly in hematological malignancies, to detect gene fusion events. For example, it can identify the fusion of the CRLF2 gene in Ph-like B-ALL.
Mutation and splicing variationdetection: Targeted RNA-seq is capable of detecting point mutations, insertions or deletions, and complex splicing variations.
Phenotype-specific expression analysis: By analyzing the RNA expression patterns in specific cell types or tissues, targeted RNA - seq contributes to uncovering the molecular mechanisms underlying diseases.
It is important to note that while targeted RNA - seq is a powerful tool for highly sensitive and specific RNA analysis in specific research areas, its application should be tailored to specific research needs. Combining it with other technologies, such as full - transcriptome RNA - seq, can yield more comprehensive results.
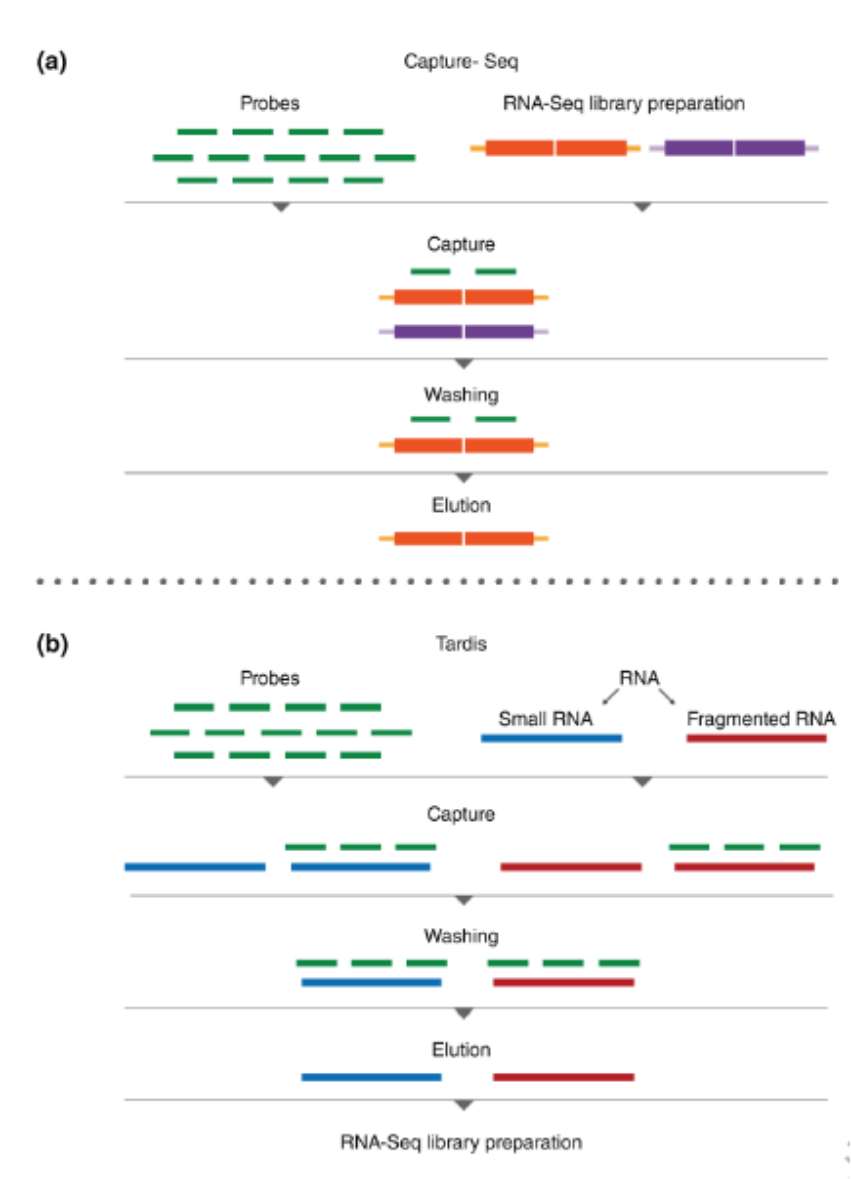 Targeted RNA-Seq by target capture (Hrdlickova et al., 2016)
Targeted RNA-Seq by target capture (Hrdlickova et al., 2016)
Services you may interested in
Learn More
The Principle of Targeted RNA-seq
Targeted RNA sequencing is a transcriptome analysis method based on high-throughput sequencing technology, which aims to deeply cover and accurately analyze the gene or RNA region of interest through specific design and experimental strategies. Its basic principle is as follows.
Selection of target region and probe design: First, it is necessary to identify the RNA target region to be sequenced. These regions are usually gene transcripts, non-coding RNA, etc. related to specific biological problems or diseases. According to the nucleotide sequence of the target region, a specific probe complementary to it is designed. Probes are generally single-stranded DNA or RNA molecules, and the length usually varies from tens to hundreds of base pairs. They can specifically bind to the target RNA sequence. In order to facilitate the subsequent operation, the probe may be marked with special labels such as biotin.
RNA extraction and fragmentation: Total RNA is extracted from biological samples (such as cells and tissues) to ensure that the extracted RNA has high integrity and purity. Then, physical methods (such as ultrasonic treatment) or enzymatic methods (such as ribonuclease) are used to cut RNA into fragments of appropriate size, generally about 100-500 bases in length, so as to facilitate subsequent hybridization and sequencing reactions.
Hybridization capture: hybridize the fragmented RNA with the designed probe. Under appropriate temperature and salt concentration, the probe will specifically bind to the target RNA fragment with complementary sequence to form RNA-probe hybrid. Non-target RNA fragments are still in a free state without binding to the probe. In order to improve the efficiency and specificity of hybridization, some reagents to promote hybridization, such as formamide, are usually added to the hybridization system.
Enrichment: RNA-probe hybrids are separated from the mixed system by using special labels labeled on probes and corresponding capture techniques. For example, if the probe is labeled with biotin, streptavidin magnetic beads can be used for capture. Streptavidin has a strong affinity with biotin, which can specifically bind to probe-RNA hybrid labeled with biotin, and then the magnetic beads and their bound target RNA fragments are separated by magnetic force to enrich the target RNA and remove a large number of non-target RNA.
Library construction and sequencing: library construction was carried out on the captured and enriched target RNA fragments. Generally, it includes adding specific linker sequences at both ends of RNA fragments to make them combine with primers of sequencing platform. Then the library was amplified by PCR to increase the amount of target RNA for subsequent sequencing. Finally, the constructed library is loaded on the high-throughput sequencing platform for sequencing, and the sequence information of RNA is determined by detecting the signal from each base, so as to obtain the detailed sequence data of the target RNA region for further bioinformatics analysis, such as gene expression analysis and mutation detection.
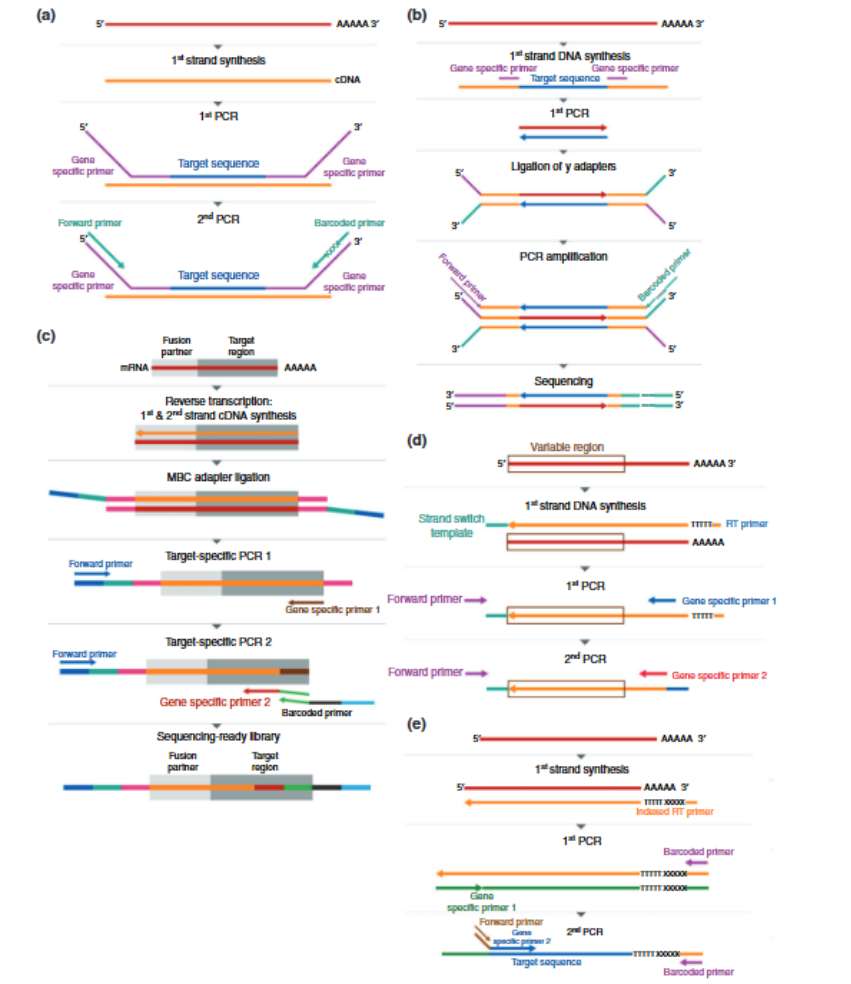 Targeted RNA-Seq by amplicon sequencing (Hrdlickova et al., 2016)
Targeted RNA-Seq by amplicon sequencing (Hrdlickova et al., 2016)
How Does Targeted RNA-seq Work
Targeted RNA sequencing generally includes sample preparation, library construction, sequencing and data analysis. The following is a detailed introduction.
Sample preparation
- Collecting samples: Select appropriate biological samples, such as blood, tissues and cells, according to the research purpose. Ensure that the sample collection process conforms to the specifications to maintain the integrity and biological activity of RNA.
- Extraction of RNA: Total RNA is extracted from the sample by appropriate methods such as TRIzol method and magnetic bead method. After extraction, the integrity of RNA is detected by agarose gel electrophoresis or bioanalyzer, and the concentration and purity of RNA are determined by spectrophotometer to ensure that it meets the requirements of subsequent experiments.
- RNA fragmentation: Physical methods such as ultrasonic treatment or enzymatic methods such as RNase are used to cut RNA into fragments of appropriate size, and the length of the fragments is generally about 100-500bp.
Library construction
- Probe hybridization: Design and synthesize a specific probe for the target RNA region, and hybridize it with the fragmented RNA under specific conditions, so that the probe can specifically bind to the complementary sequence of the target RNA.
- Capture and enrichment: The target RNA fragment bound to the probe is captured by using biotin and other labels labeled on the probe, and the unbound RNA and impurities are removed by washing and other operations to realize the enrichment of the target RNA.
- Reverse transcription: Reverse transcriptase and primers are added to the enriched target RNA, and cDNA is synthesized with RNA as a template.
- Add linker: Add specific linker sequences at both ends of cDNA to provide binding sites for subsequent PCR amplification and sequencing.
PCR amplification: The cDNA with linker is amplified by PCR reaction to increase the number of target sequences for subsequent sequencing analysis.
Sequencing
- On-line sequencing: The constructed library is loaded on high-throughput sequencing platform, such as Illumina and PacBio, and sequenced according to their respective sequencing principles and processes to generate a large number of original sequencing data.
Data analysis
- Data preprocessing: Quality control of original sequencing data, removal of low-quality sequences, linker sequences, etc. to improve the accuracy and reliability of data.
- Comparison and annotation: Compare the processed data with the reference genome or transcriptome, determine the position of each sequence in the genome, and make gene annotation on it to identify the specific gene and transcript information of the target RNA.
- Differential analysis: According to the research purpose, the differential expression of genes between different samples was analyzed to find out the differentially expressed genes, and further explore the key genes and pathways related to diseases or biological processes.
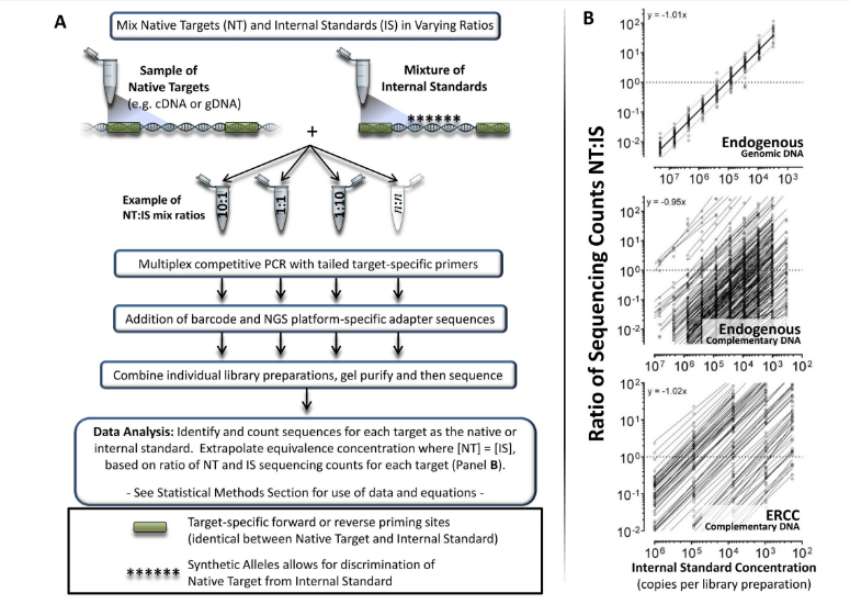 Competitive amplicon library preparation workflow and data analysis overview (Blomquist et al., 2013)
Competitive amplicon library preparation workflow and data analysis overview (Blomquist et al., 2013)
The Advantages and Disadvantages of Targeted RNA-seq
As an important sequencing technology, targeted RNA sequencing is widely used in biology and medical research. The following are its advantages and disadvantages.
Advantages
- High sensitivity and specificity: Targeted RNA sequencing can provide higher sequencing depth and sensitivity by specifically capturing the target RNA molecules, especially for the detection of low-abundance transcripts. For example, in the diagnosis of hematological malignancies, targeted RNA-seq can identify clinically relevant markers (such as LSC177 or pLSC632) and detect acquired somatic mutations.
- Cost-effectiveness: Targeted RNA sequencing reduces the sequencing cost by reducing the sequencing amount of non-target RNA. For example, the preparation method of competitive multiplex PCR amplification library can maintain consistency between different dates, library preparation and laboratories, while significantly reducing costs.
- Reduce background interference: Targeted RNA sequencing avoids the problem of nonspecific probe binding or cross-probe hybridization, thus reducing background interference. In addition, by optimizing the design, false positive signals can be reduced.
- Flexibility and adaptability: The targeted RNA sequencing platform is highly adaptable and can be integrated into Qualcomm screening pipelines and compound development to study the expression of stress proteins in specific signal pathways.
- Suitable for complex samples: Targeted RNA sequencing performs well in complex samples (such as blood and tumor tissues), and can detect fusion transcripts, gene mutations and exon jumps.
Disadvantages
- Lack of design complexity: Targeted RNA sequencing requires carefully designed primers and probes to ensure the specific capture of the target RNA. This may require professional knowledge and experience
- Limited coverage: Targeted RNA sequencing usually only sequences specific RNA molecules or gene regions, so it can't fully cover the whole transcription group. This may result in the omission of genes or transcripts that are not captured by the target.
- Technical limitation: Targeted RNA sequencing may be affected by the deviation of PCR amplification, especially in the preparation of multiplex PCR amplification library, different targets may be biased due to the difference of amplification efficiency. In addition, some techniques (such as molecular barcode noise reduction) are rarely used in targeted sequencing because they may introduce non-specific interference.
- Complex data interpretation: the data interpretation of targeted RNA sequencing needs to be combined with biological background knowledge to distinguish real signals from potential technical noise. For example, when detecting newborn RNA, it may be affected by RNA degradation during immunoprecipitation.
- Limited scope of application: Targeted RNA sequencing is more suitable for studying specific biological problems or disease mechanisms than comprehensive genome analysis. For example, in microbial analysis, whole genome sequencing method may be more suitable for species abundance estimation.
Targeted RNA sequencing is an efficient, economical and sensitive technology, which is suitable for the research of specific RNA molecules and disease diagnosis. However, its design complexity, limited coverage and technical limitations are the main challenges to be overcome.
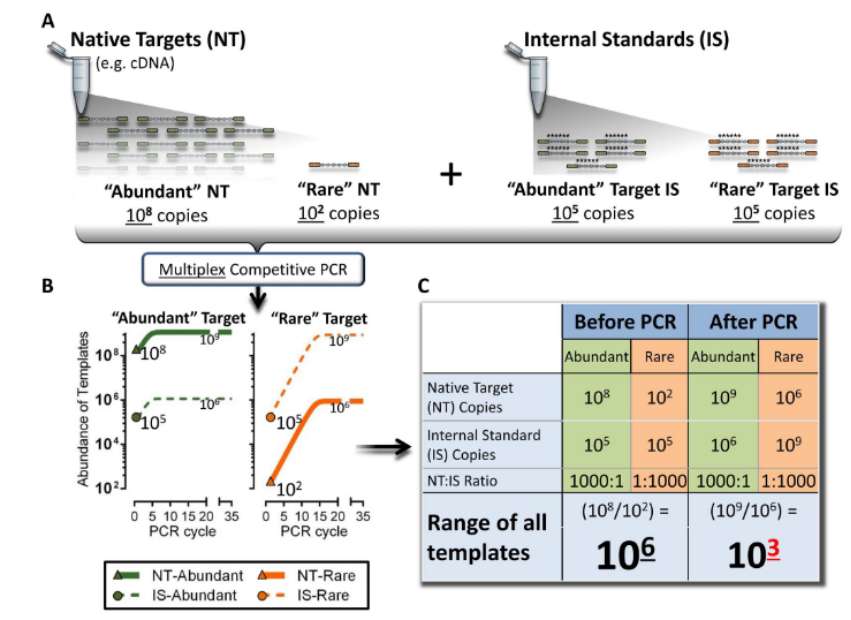 Schematic depiction of how competitive amplicon library preparation reduces oversampling (Blomquist et al., 2013)
Schematic depiction of how competitive amplicon library preparation reduces oversampling (Blomquist et al., 2013)
Applications of Targeted RNA-seq
Targeted RNA sequencing is a high-precision transcriptome analysis technology, which can efficiently detect and quantify the genes of interest by enriching or amplifying RNA in specific genomic regions. This technology has been widely used in many fields, including cancer research, drug development, neurogenomics and clinical diagnosis.
Cancer research
Targeted RNA sequencing has important applications in cancer research, especially in tumor gene fusion detection, gene mutation and expression profile analysis.
- Detection of tumor gene fusion: Targeted RNA sequencing can identify known and unknown gene fusion partners, especially in fixed tissue samples (such as FFPE). This method can find rare fusion genes caused by recessive translocation or microdeletion, thus providing new targets for cancer treatment.
- Analysis of cancer heterogeneity: Targeted RNA sequencing technology can reveal the heterogeneity within the tumor and help to understand the complexity and treatment response of the tumor.
- Early cancer detection: Targeted RNA sequencing improves the sensitivity and accuracy of early cancer detection by enriching long-chain noncoding RNA(lncRNA).
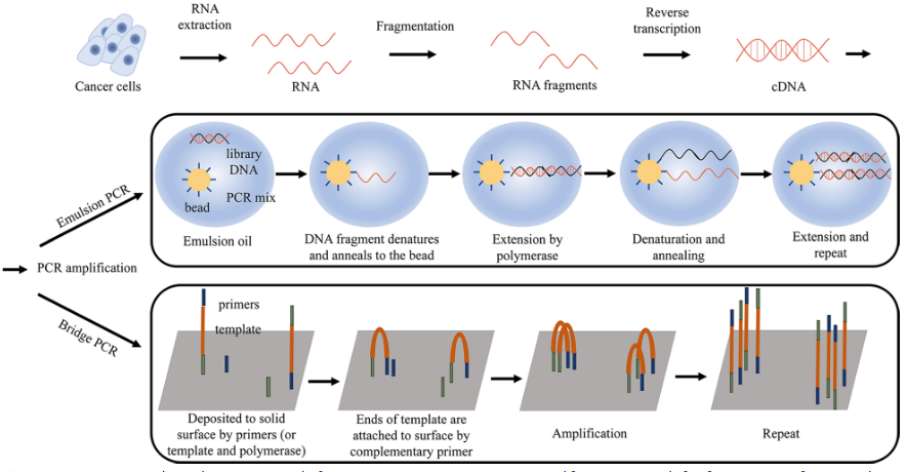 RNA extraction and template preparation in cancer (Hong et al., 2020)
RNA extraction and template preparation in cancer (Hong et al., 2020)
Drug development
Targeted RNA sequencing plays an important role in drug development. It can accurately determine specific RNA regions, help screen drug targets, evaluate drug efficacy by analyzing RNA changes before and after drug action, and also find the characteristics of drug-resistant related RNA, which provides key basis for optimizing drugs and predicting drug resistance risks, and accelerates the research and development process of safe and effective drugs.
- Gene expression regulation: Through targeted RNA sequencing, researchers can analyze the gene expression changes of cells after drug treatment and identify potential drug targets.
- Disease model construction: Using Qualcomm RNA sequencing technology, we can establish a tumor heterogeneity model from patients for drug response prediction.
Neurogenomics
Targeted RNA sequencing is widely used in neurogenome research. It can accurately focus on the transcription of nerve-related genes, reveal the gene expression differences and alternative splicing abnormalities in nerve development and neurodegenerative diseases such as Alzheimer's disease by analyzing specific RNA sequences, and help to analyze the mechanism of nerve diseases and explore potential therapeutic targets.
- Neural development research: By analyzing the transcriptome of nerve cells, we can reveal the gene expression pattern in the process of neural development.
- Study on the mechanism of neurological diseases: Targeted RNA sequencing can help identify abnormal gene expression related to neurological diseases and provide clues for the diagnosis and treatment of diseases.
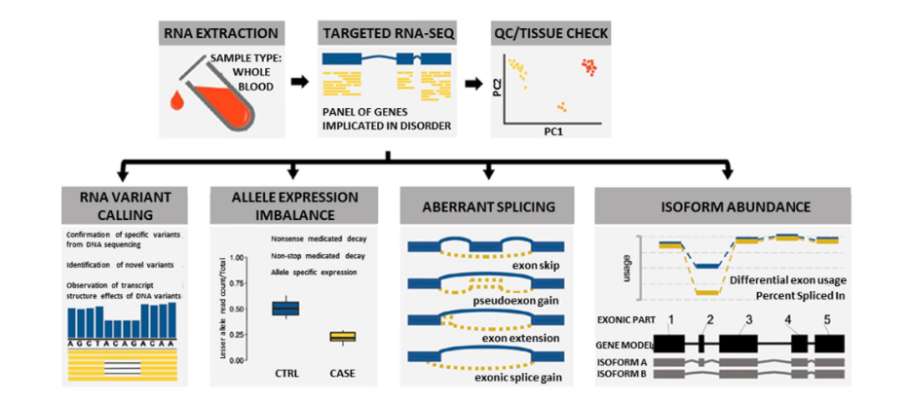 Overview of approach used to analyze RNA to aid in the diagnosis of dysferlinopathy (Rufibach et al., 2023)
Overview of approach used to analyze RNA to aid in the diagnosis of dysferlinopathy (Rufibach et al., 2023)
Clinical diagnosis
Targeted RNA sequencing is widely used in clinical diagnosis, which can accurately detect tumor-related gene transcripts and assist early screening, typing and prognosis judgment of cancer. Can identify the pathogen RNA and help the rapid diagnosis of infectious diseases; It can also be used to detect RNA variation related to genetic diseases, and provide key basis for clinical diagnosis, treatment plan formulation and disease monitoring.
- Detection of fusion genes: Through targeted RNA sequencing, fusion genes in hematological malignancies, such as LSC17 or P63LSC2, can be detected, thus providing support for clinical diagnosis.
- Diagnosis of rare diseases: Targeted RNA sequencing can be used to identify the pathogenic genes of rare diseases and help diagnosis and treatment by analyzing the expression patterns of specific genes.
Case Study of Targeted RNA-seq in Gene Fusion
Fusion genes can lead to abnormal sequences or functional protein, or the imbalance of some genes' expression, and then lead to or promote the occurrence of tumors. It is reported that the incidence of human cancer caused by fusion genes is about 20%. However, the prevalence of fusion genes varies greatly in different cancers, and many fusion genes are specific to specific cancer subtypes. Therefore, rapid and accurate identification of fusion genes can qualitatively and hierarchically diagnose cancer.
RNA was extracted from K562 and RDES cell lines, and compared with conventional RNA-seq by targeted RNA-seq. The results showed that the ERCC abundance of targeted RNA-seq sequencing was 59 times (blood panel) and 33 times (solid tumor panel) than that of conventional RNA-seq sequencing, and the minimum input of targeted capture sequencing was 3pM. This shows that both Panel in RNA targeted sequencing have strong capture ability.
The researchers evaluated the reliability of the fusion gene detected by two panel. The results showed that the transcript coverage uniformity of Panel in hematological tumor and solid tumor was good, and the coverage of known splicing sites was 77.8% and 84.6% respectively. This shows that both Panel in RNA targeted capture sequencing have the ability to detect most fusion genes.
The study also compared the detection of low-copy fusion genes between targeted RNA-seq and conventional RNA-seq. The results showed that there was no significant difference between the two methods in detecting BCR-ABL1 (8-24 DNA copy) in K562 cell line. However, when EWSR1-FLI1 (single DNA copy) was detected in RDES cell line, conventional RNA-seq sequencing was not covered by reads at the beginning of the fusion site. This shows that RNA targeted capture sequencing has obvious advantages over conventional RNA-seq sequencing in detecting low abundance fusion genes.
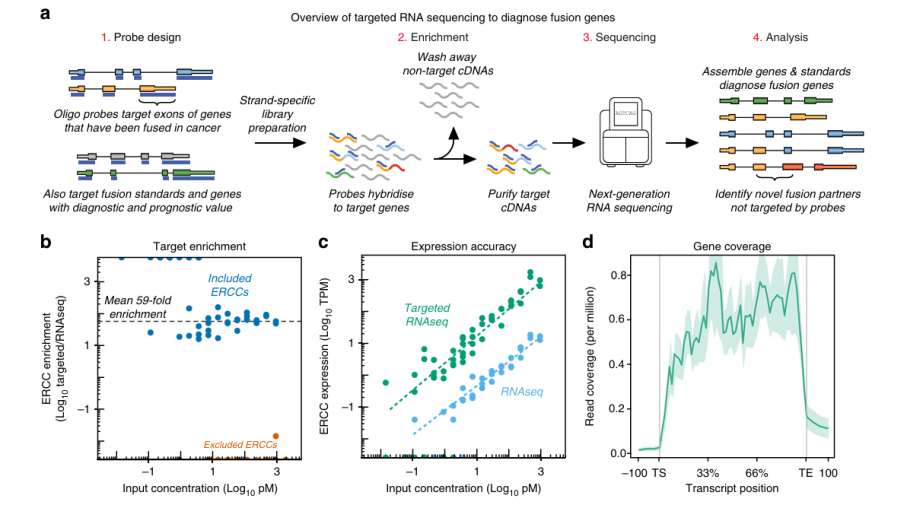 Overview of targeted RNA-seq and panel validation (Heyer et al., 2019)
Overview of targeted RNA-seq and panel validation (Heyer et al., 2019)
Conclusion
Targeted RNA sequencing, as a powerful means to accurately study specific RNA, has a unique principle. Firstly, it designs complementary probes according to the target RNA region, which can specifically bind to the target RNA like "precise navigation". Sample RNA is extracted, fragmented and hybridized with the probe, and then the target RNA-probe hybrid is captured and enriched by the marker. Then, the library was constructed and sequenced to realize the deep sequencing of the target RNA region. Its advantages are obvious. Compared with complete transcriptome sequencing, it can focus more on key regions, with lower cost and higher sequencing depth, which is convenient for accurate detection of low-abundance RNA. However, it also has limitations, only for known target areas, and may miss new transcripts or variations.
In the application field, targeted RNA sequencing is widely used. In scientific research, it helps to explore gene functions, and reveals its role in biological processes by accurately detecting the expression of specific genes. In the medical field, it can be used for disease diagnosis and prognosis evaluation, such as targeted sequencing of tumor-related genes, assisting early diagnosis and disease monitoring of tumors. It can also be used in drug research and development. By monitoring the changes of target RNA under the action of drugs, the efficacy and potential toxicity can be evaluated, providing key data support for new drug development.
References:
- Hrdlickova R, Toloue M, Tian B. "RNA-Seq methods for transcriptome analysis." Wiley Interdiscip Rev RNA. 2017 8(1):10.1002/wrna.1364 https://doi.org/10.1002/wrna.1364
- Blomquist TM, Crawford E., et al. "Targeted RNA-sequencing with competitive multiplex-PCR amplicon libraries." PLoS One. 2013 8(11):e79120 https://doi.org/10.1371/journal.pone.0079120
- Hong Mingye, Tao Shuang., et al. "RNA sequencing: new technologies and applications in cancer research." Journal of Hematology & Oncology. 2020: 166 https://doi.org/10.1186/s13045-020-01005-x
- Rufibach L, Berger K., et al. "Utilization of Targeted RNA-Seq for the Resolution of Variant Pathogenicity and Enhancement of Diagnostic Yield in Dysferlinopathy." J Pers Med. 2023 13(3):520 https://doi.org/10.3390/jpm13030520
- Heyer, E.E, Deveson, I.W, Wooi, D., et al. "Diagnosis of fusion genes using targeted RNA sequencing." Nat Commun 10 1388 (2019) https://doi.org/10.1038/s41467-019-09374-9
- Athanasopoulou K, Daneva GN., et al. "The Transition from Cancer "omics" to "epi-omics" through Next- and Third-Generation Sequencing." Life (Basel). 2022 12(12):2010 https://doi.org/10.3390/life12122010
- Haile S, Corbett RD., et al. "Evaluation of protocols for rRNA depletion-based RNA sequencing of nanogram inputs of mammalian total RNA." PLoS One. 2019 14(10):e0224578 https://doi.org/10.1371/journal.pone.0224578
- Yap KL, Furtado LV., et al. "Diagnostic evaluation of RNA sequencing for the detection of genetic abnormalities associated with Ph-like acute lymphoblastic leukemia (ALL)." Leuk Lymphoma. 2017 58(4):950-958 https://doi.org/10.1080/10428194.2016.1219902


 Sample Submission Guidelines
Sample Submission Guidelines
